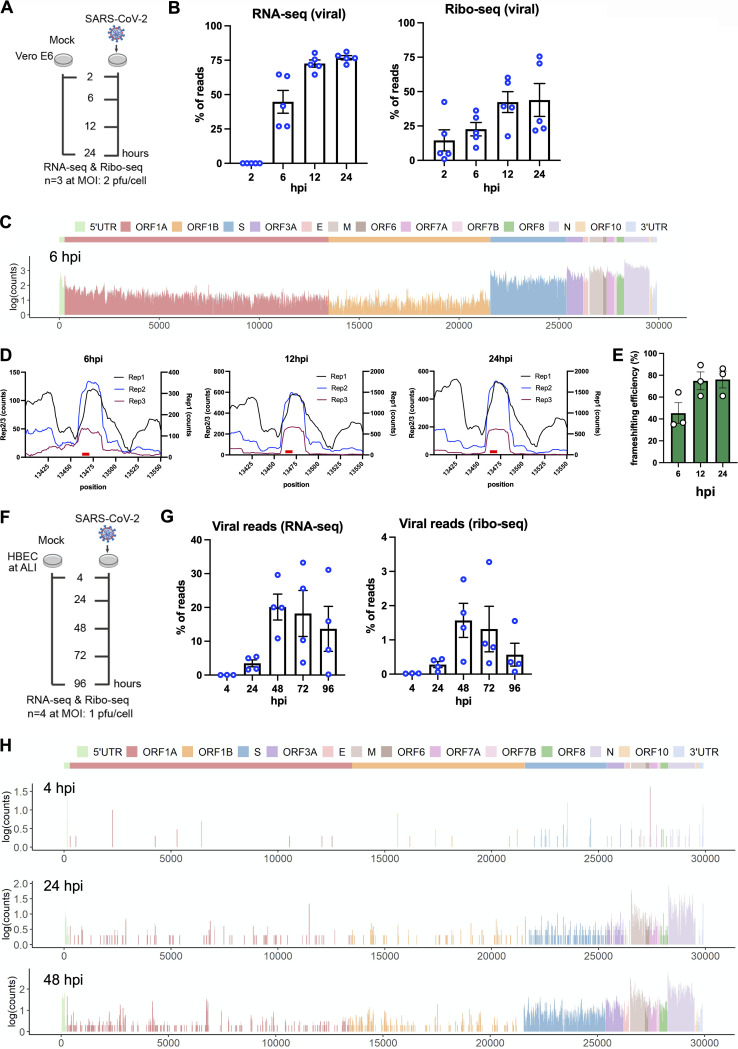
FIG 1.
Ribo-seq reveals the translational program of SARS-CoV-2. (A) Schematic diagram of ribo-seq and RNA-seq experiments conducted in this study. Vero E6 cells were infected at 2 PFU/cell, and cells were processed for RNA-seq and ribo-seq at 2, 6, 12, and 24 hpi. (B) Percentages of RNA-seq and Ribo-seq reads uniquely mapping to SARS-CoV-2 and cellular transcripts at the indicated time points postinfection. Individual data points indicate independent biological replicates. (C) Ribo-seq counts (log10) along the viral genome at 6 hpi (see Fig. S3). The schematic diagram of SARS2 genome features shown at the top is colinear (see Table S3). (D) Ribo-seq read counts within the frameshifting site across three independent replicates at 6, 12, and 24 hpi. (E) SARS-CoV-2 frameshifting efficiency as determined by comparing the average read densities between ORF1a and ORF1b regions across three independent replicates and various time points postinfection. (F) Schematic diagram of ribo-seq and RNA-seq experiments conducted in this study. HBECs grown at ALI were infected at 1 PFU/cell and cells were processed for RNA-seq and Ribo-seq at 4, 24, 48, 72 and 96 hpi. (G) Percentage of RNA-seq and Ribo-seq reads uniquely mapping to SARS-CoV-2 and cellular transcripts at the indicated time points postinfection. Note that infection in this system progresses slower than in Vero E6 cells and a relatively small percentage of cells are infected at 24 and 48 hpi, as exemplified in Fig. S6. Individual data points indicate independent biological replicates. (H) Ribo-seq counts (log10) along the viral genome across various time points. The schematic diagram of SARS2 genome features shown at the top is colinear (Fig. S7 and Table S10).