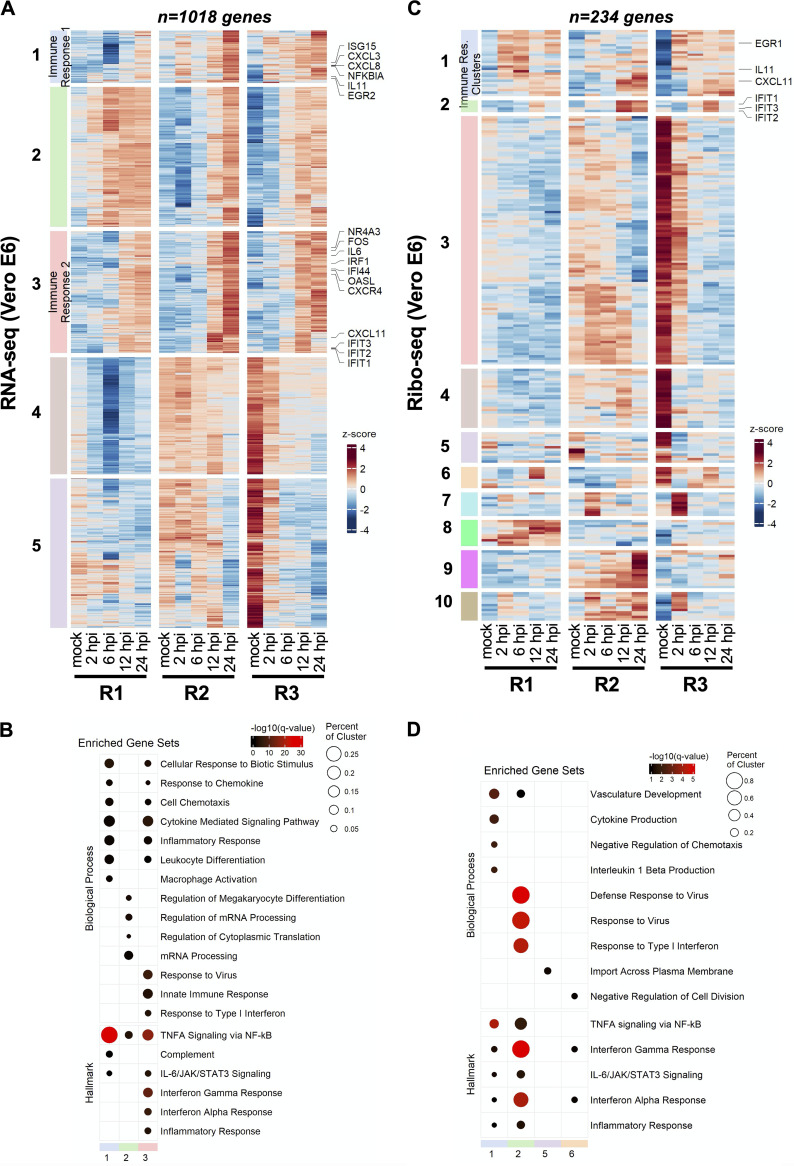
FIG 2.
SARS-CoV-2 infection induces translational repression of innate immune genes. Vero E6 cells infected at 2 PFU/cell as detailed in Fig. 1 were analyzed for differential expression of host genes by RNA-seq (A and B) and ribo-seq (C and D). (A and C) Hierarchical clustering of DEGs after infection. Genes were filtered for an absolute log2 fold change of >2 and an adjusted q value of <0.05 at any time point. (B and D) Hypergeometric enrichment analysis from Hallmark and Gene Ontology databases for each individual cluster in 2A and 2C. Color represents significance (q value); size indicates the percentage of the cluster represented in the pathway. (See Tables S11 and S12.)