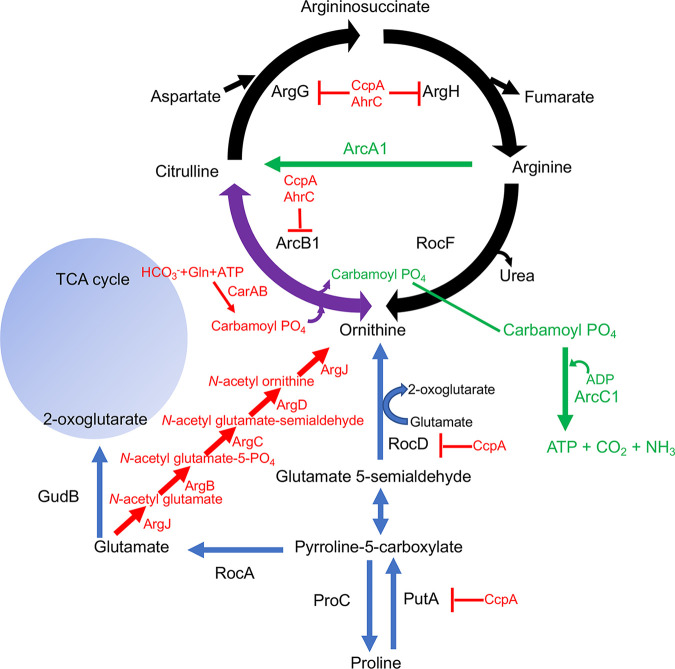
FIG 1.
Arginine metabolic pathways in Staphylococcus aureus. Proline serves as a substrate for arginine biosynthesis via PutA, RocD, ArcB1, ArgG, and ArgH. The transcription of putA, rocD, arcB1, and argGH are repressed by CcpA, whereas arcB1 and argGH are repressed by AhrC and CcpA. The canonical arginine biosynthetic pathway (in red) is repressed under the conditions studied in this article. The arginine deiminase pathway (shown in green) generates ATP, CO2, and NH3. All three pathways utilize ornithine carbamoyltransferase (ArcB1) (shown in purple). TCA, tricarboxylic acid.