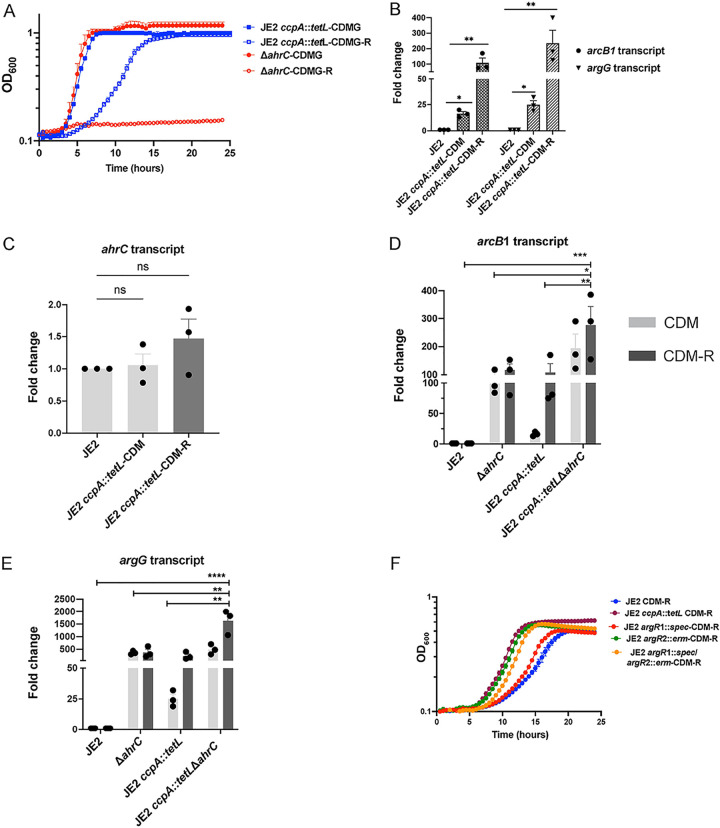
FIG 6.
CcpA represses argGH and arcB1 transcription during growth in CDM-R. (A) Growth analysis of JE2 ΔahrC and JE2 ccpA::tetL in CDMG and CDMG-R. Data represent results from three technical replicates per strain. Data are represented as means ± SEM. (B) qRT-PCR assessing the transcripts of arcB1 and argG in JE2 ccpA::tetL in both CDM (light gray) and CDM-R (dark gray). Statistical significance was assessed using one-way ANOVA followed by Dunnett’s posttest. Asterisks indicate significant differences between WT JE2 and JE2 ccpA::tetL (grown in CDM and CDM-R). (C) Expression of ahrC in JE2 ccpA::tetL in CDM and CDM-R compared to JE2. Statistical significance was assessed using one-way ANOVA followed by Dunnett’s posttest. (D and E) Expression of arcB1 (D) and argG (E) in JE2 ccpA::tetL ΔahrC in both CDM (light gray) and CDM-R (dark gray). Data are representative of results from three independent biological replicates performed with two technical replicates in each experiment. Error bars show means ± SEM (n = 3). Statistical significance was assessed using one-way ANOVA followed by Dunnett’s posttest and Tukey’s posttest. Significance was assessed between JE2 ccpA::tetL ΔahrC (grown in CDM-R) and JE2 (grown in CDM), JE2 ccpA::tetL (grown in CDM-R), and JE2 ΔahrC (grown in CDM-R). (F) Growth of JE2, JE2 argR1, JE2 argR2, JE2 argR1 argR2, and JE2 ccpA in CDM-R. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.001; ns, no significance.