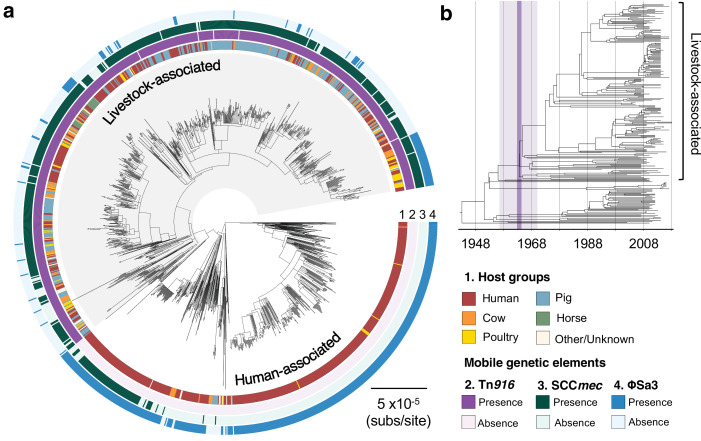
Figure 1. The transition to livestock association in the 1960s was accompanied by changes in the frequencies of three mobile genetic elements (MGEs).
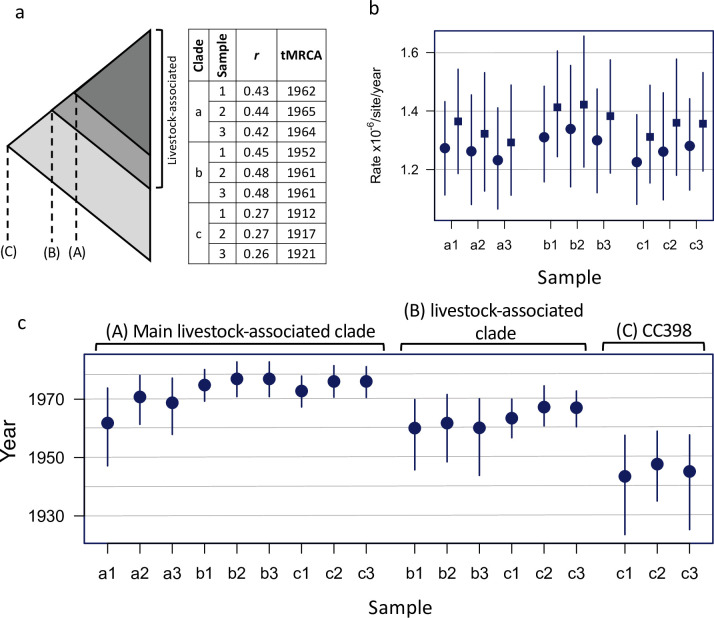
(a) A maximum likelihood phylogeny of 1180 isolates of CC398, rooted using an outgroup from ST291. Grey shading indicates the livestock-associated clade. Outer rings describe (1) the host groups isolates were sampled from, and the presence of three MGEs: (2) a Tn916 transposon carrying tetM, (3) a SCCmec carrying mecA, and (4) a φSa3 prophage carrying a human immune evasion gene cluster. (b) A dated phylogeny of a sample of 250 CC398 isolates that shows livestock-associated CC398 originated around 1964 (95% HPD: 1957–1970).