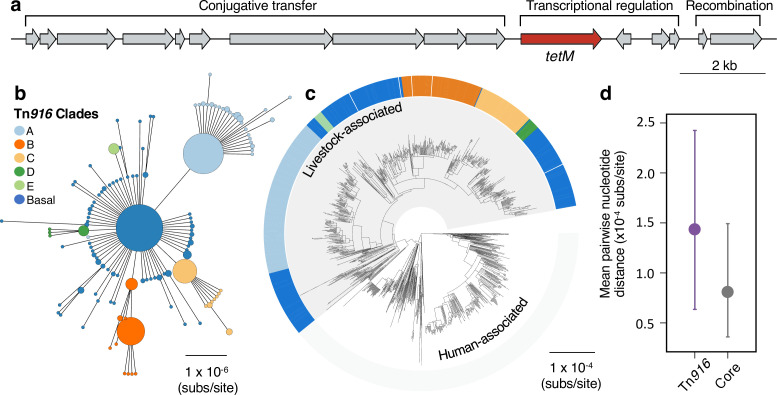
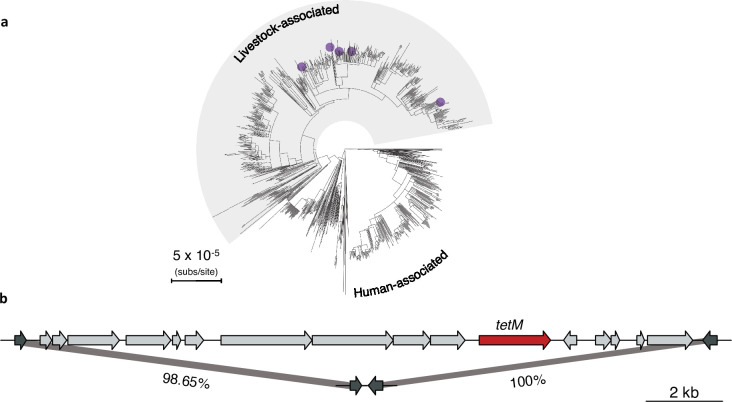
Figure 2. A Tn916 transposon carrying tetM has been stably maintained by livestock-associated CC398 since its origin.
(a) A gene map of the Tn916 transposon in CC398 (based on reference genome 1_1439), with annotations based on previous studies (de Vries et al., 2009; Roberts and Mullany, 2009). (b) A minimum-spanning tree of the element based on a concatenated alignment of all genes shown in (a). Points represent groups of identical elements, with point size correlated with number of elements on a log scale, and colours representing well-supported clades (>70 bootstrap support in a maximum likelihood phylogeny) that include >10 elements (smaller clades are incorporated into their basal clade). (c) These clades are annotated onto the CC398 phylogeny as an external ring. (d) Mean pairwise nucleotide distance between isolates carrying the Tn916 transposon based on genes in the Tn916 transposon and core genes, using bootstrapping to estimate error (see Materials and methods for details).