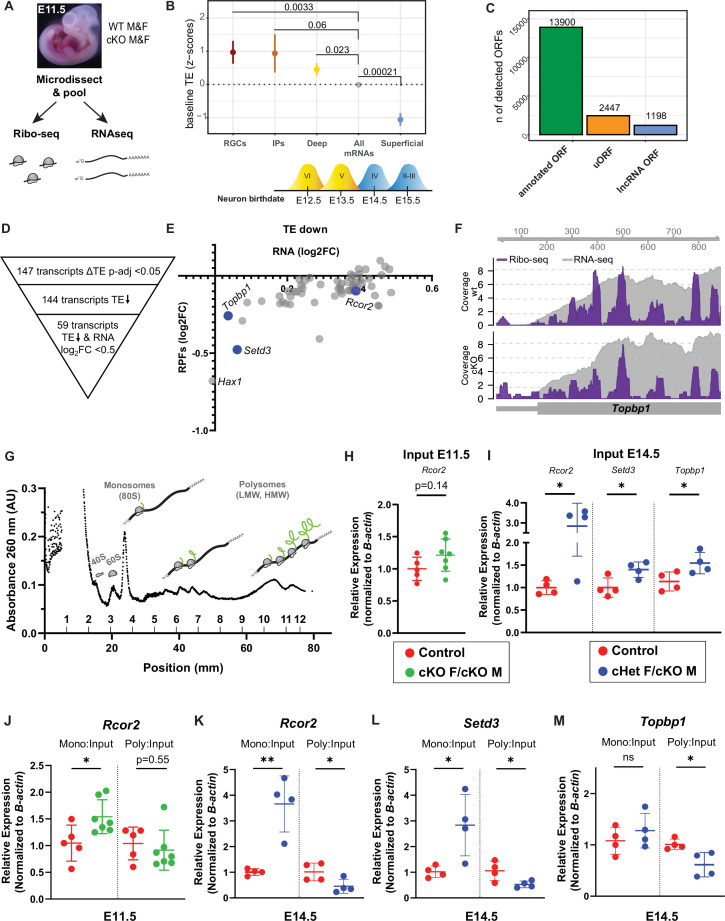
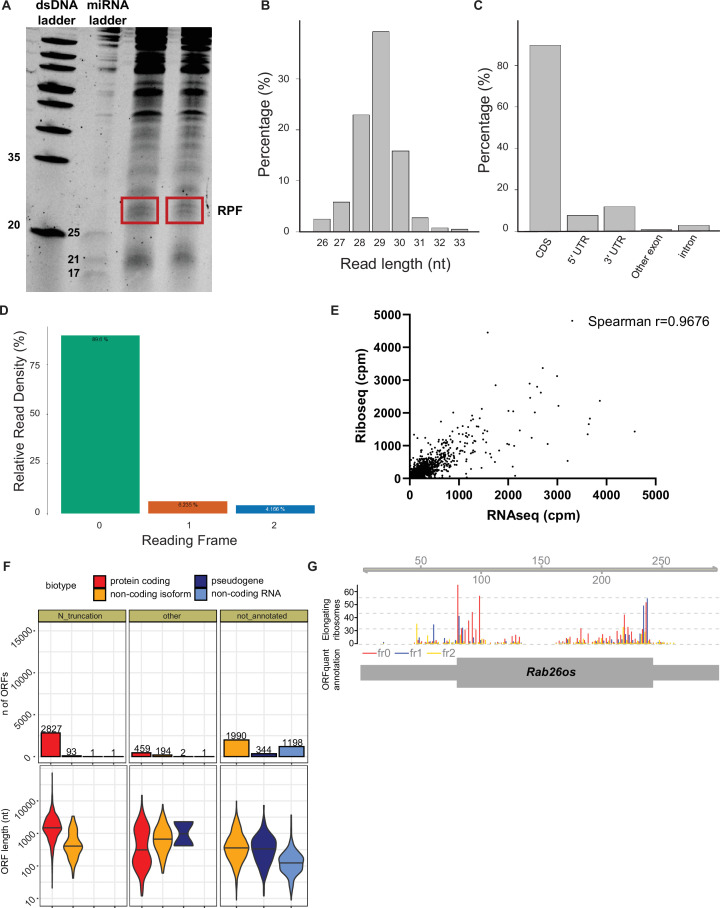
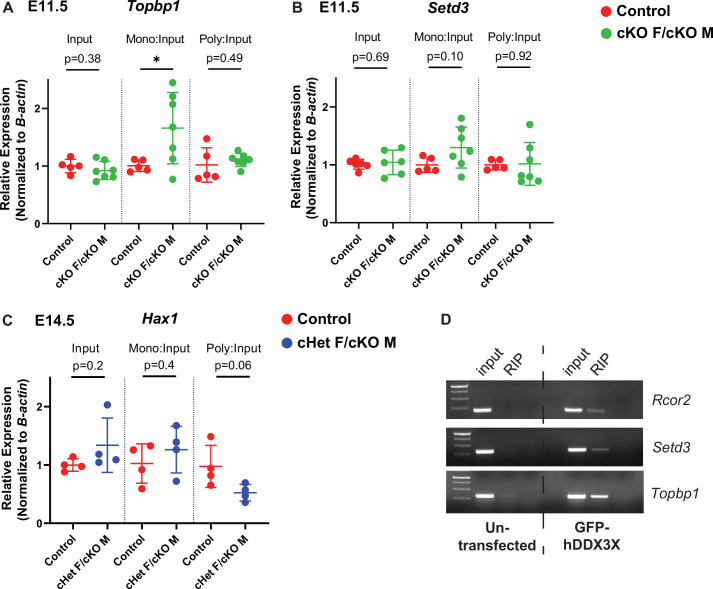
Figure 6. Ribosome Profiling in embryonic brains uncovers the E11.5 translatome including DDX3X-dependent translation targets.
(A) Experimental paradigm for Ribo-seq and RNAseq of E11.5 cortices from control and cKO mice. n=3/sex/condition with four embryos pooled per n. (B) TE of transcripts enriched in RGCs, IPs, deep layer neurons (VI-V) and superficial layer neurons (IV-II) relative to all other mRNAs (TPM >10). Birthdates for laminar layers are indicated below. See Supplementary file 3 for exact transcripts. (C) ORFquant analysis of wildtype Ribo-seq data showing identification of annotated ORFs and uORFs in protein-coding and non-coding isoforms. (D) Schematic illustrating how DDX3X-dependent targets were prioritized. (E) Scatter plot of RPFs log2FC versus RNA log2FC for 59 DDX3X-dependent targets with significantly lower TE. Putative Ribo-seq targets selected for validation are highlighted in blue. (F) IGV screenshots illustrating RNAseq reads (gray) and RPFs (Ribo-seq; purple) for Topbp1 in cKO mice relative to control. (G) Representative trace from polysome fractionation of E14.5 cortical lysate. (H–M) RT-qPCR quantification of mRNA levels for Ribo-seq candidates in input samples at E11.5 (H) and at E14.5 (I), and monosome and polysome fractions at E11.5 (J) and E14.5 (K–M). n=5–7/condition (H, J) and 4/condition (I, K–M) with two embryos pooled per n. Error bars, S.D. *p<0.05, **p<0.01. Two-sided Wilcoxon test (B), Student’s unpaired, two-tailed t-test (H–M).