Fig. 2.
ORF8 escapes ER degradation by forming mixed disulfide complexes.
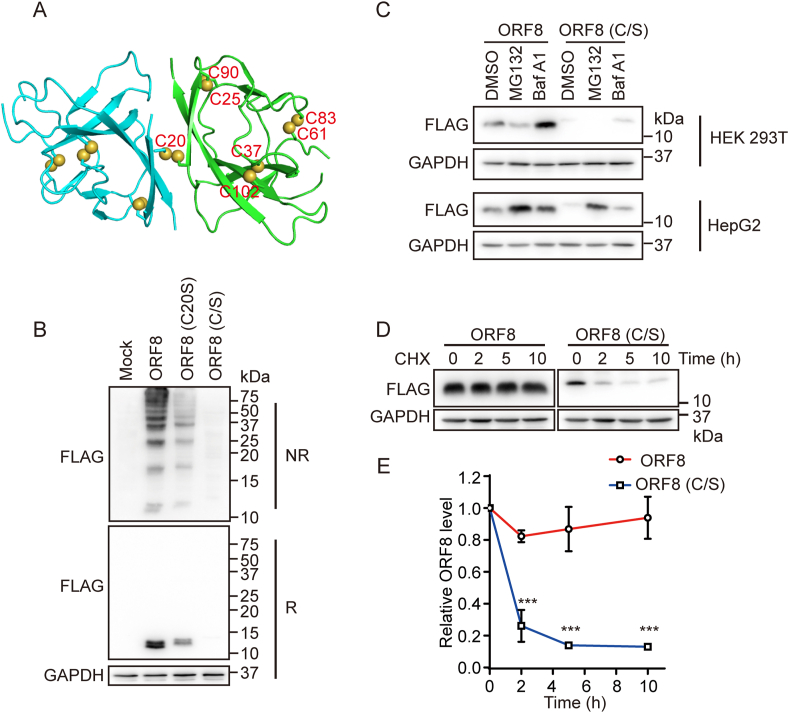
(A) Crystal structure of SARS-CoV-2 ORF8 with two monomers shown in cyan and green (PDB: 7JTL). Cysteine residues were shown as spheres.
(B) Immunoblotting analysis of the redox state of ORF8 in HEK 293T cells expressing ORF8-FLAG or the mutants. Cell lysates were separated by SDS-PAGE under either reducing (R) or nonreducing (NR) conditions.
(C) Immunoblotting of ORF8 in HEK 293T and HepG2 cells expressing ORF8-FLAG or the C/S mutant treated with 100 nM Bafilomycin A1 (BafA1) or 8 μM MG132 for 12 h.
(D) Immunoblotting of ORF8 in HEK 293T cells expressing ORF8-FLAG and the C/S mutant treated with 100 nM BafA1 for 15 h and then followed by treatment with 25 μg/ml cycloheximide (CHX) for indicated time points.
(E) The relative ratios of ORF8/GAPDH normalized to that at zero time point were quantified. The band intensities ORF8 and GAPDH in (D) were analyzed using the ImageJ software. The data were shown as mean ± SEM from three independent experiments; ***p < 0.001 (two-tailed Student's t-test). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)