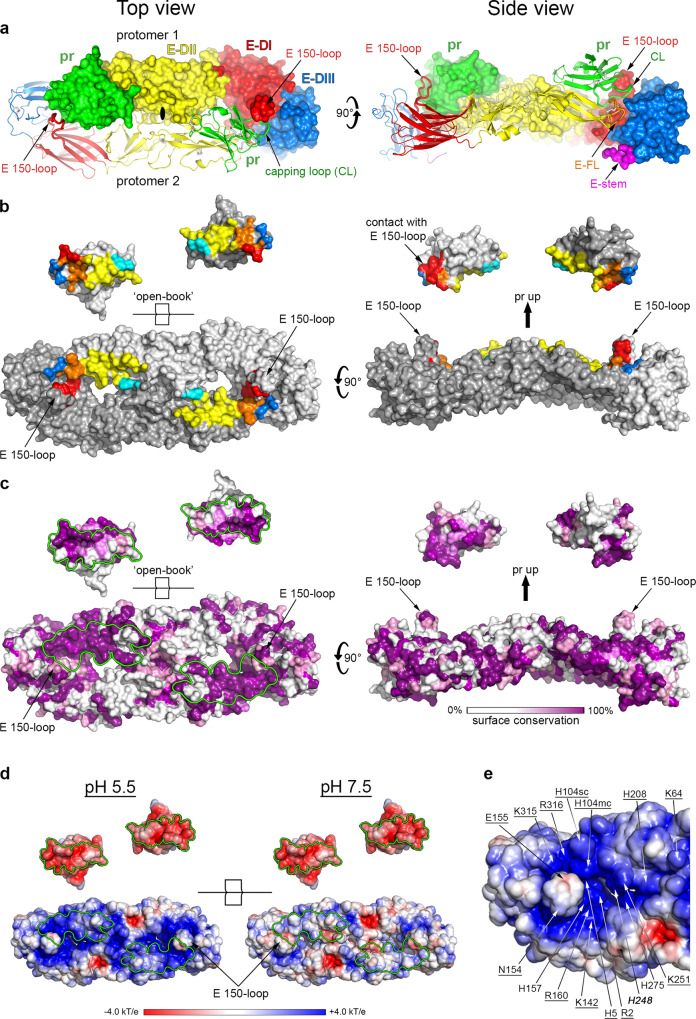
Fig. 2. X-ray structure of the (pr/sE)2 dimer.
a The (pr/sE)2 dimer shown in two orthogonal views, with E color-coded by domains as indicated (E domain I red, II yellow, III blue, stem magenta and fusion loop (E-FL) in orange). pr is colored green. One pr/sE protomer is shown in ribbons and the other in surface representation, with a central solid black oval in the top view marking the crystallographic 2-fold symmetry axis. Elements such as the E fusion loop (FL), E 150-loop, E stem (residues 396–400 downstream domain III) and pr capping loop (CL) discussed in the text are labeled. b pr footprint on the sE dimer surface. The two protomers are shown in two shades of grey in an open-book representation (left panel), and with the pr subunits shifted up in side view (right panel). The buried surfaces in the complex are colored according to the E domains involved, as in (a), except that the inter-protomer contacts with domain II are colored cyan instead of yellow. c The same surface as in (b) heat colored as indicated in the bar underneath to highlight amino acid conservation at the pr and sE dimer surface across tick-borne flaviviruses infecting vertebrates (from the alignment shown in Supplementary Fig. 2). The buried surfaces of E and pr in the complex are outlined in green in the left panel. Note that the interaction surface is the most conserved region of both, pr and E. d Same view as in the left panels in (b) and (c), colored according to the electrostatic surface potential (as indicated in the color-code bar) computed at pH 5.5 (left panel) and at pH 7.5 (right panel). The interaction surfaces are outlined in green. e Closeup of the electrostatic potential of the sE dimer surface of interaction with pr at pH 5.5, showing numerous charged residues (labeled). Residues conserved in tick-borne flaviviruses are underlined, and those conserved in all flavivirus are in bold and italic. The glycosylated Asn154 is also labeled.