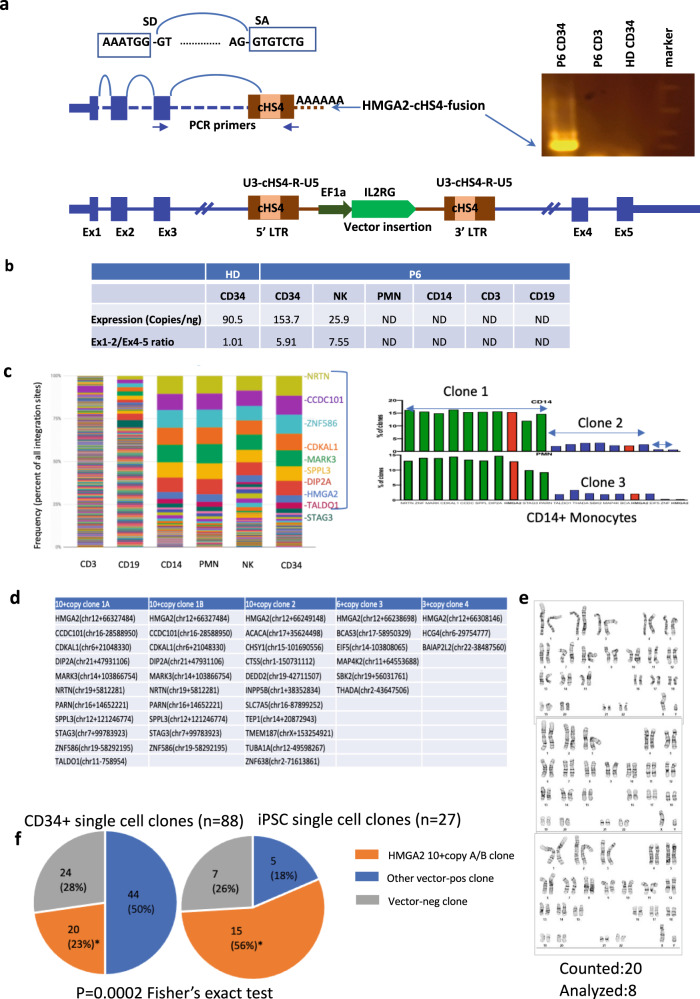
Fig. 3. Characterization of HMGA2-cHS4 fusion transcripts in single-cell CD34+ clones and derived-iPSCs clones with potential synergy in multi-insert clones.
a Illustration of lentivector insertion in the 3rd intron of HMGA2 gene in the bottom of the panel. Fusion transcript was generated by HMGA2 exon1-exon2-exon3 and spliced into the cHS4 SA in the 5LTR of the vector. Splicing Donor (SD) of HMGA2 exon3 and splicing acceptor (SA) of cHS4 sequences are shown on the top. PCR primers used to amplify the fusion transcript from P6 CD34 cells are located on HMGA2 exon3 and vector LTR. Amplified fusion transcript from RTPCR is shown in the right. b Quantification of HMGA2 transcript in different blood lineages in P6 and the 5′/3′ ratio, both determined by ddPCR as described in methods. HMGA2 is expressed in healthy donor (HD) CD34 cells, P6 CD34 cells, as well as P6 NK cells. It is not detected (ND) in P6 PMN, CD14, CD3, CD19 cells. The elevated 5′/3′ ratio suggest accumulation of truncated transcript relative to full-length transcript. c P6 Clonality at 30 m based on VISA. d Single-cell colony assay identified clones with multicopy transgenes. e Karyotype in iPSCs derived from P6 peripheral blood CD34+ cells. f HMGA2-10+ copy clones are the most abundant clones in peripheral CD34+ cells in PT6 and this clone is significantly enriched in iPSC cells derived from CD34+ cells (*p = 0.0002, Fisher’s exact test). Source data are provided as a Source Data file.