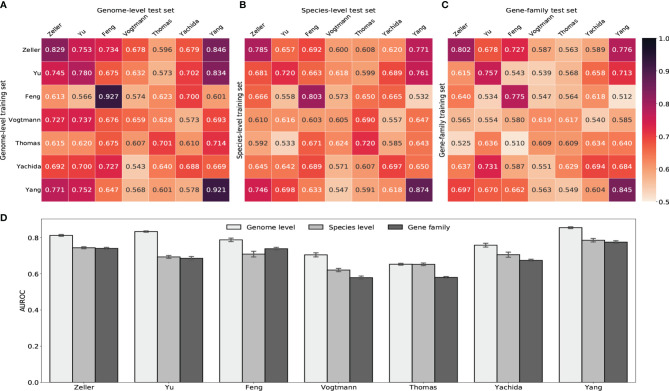
Figure 5.
Prediction performances of random forest classifiers based on gut viral abundance. (A) Within and cross study AUROC matrix obtained by using GPD genome-level abundance. The diagonal refers to results of cross validation within each dataset. Off-diagonal values refer to prediction results trained on the study of each row and tested on the study of each column. (B) Within and cross study AUROC matrix obtained by using species-level abundance. See Supplementary Figures S12A, B for genus-level and family-level AUROC. (C) Within and cross study AUROC matrix obtained by using gene-family abundance. See Supplementary Figure S12C for pathway AUROC. (D) LODO results with the x axis indicating the study left out as the validation set and other studies combined as the training set.