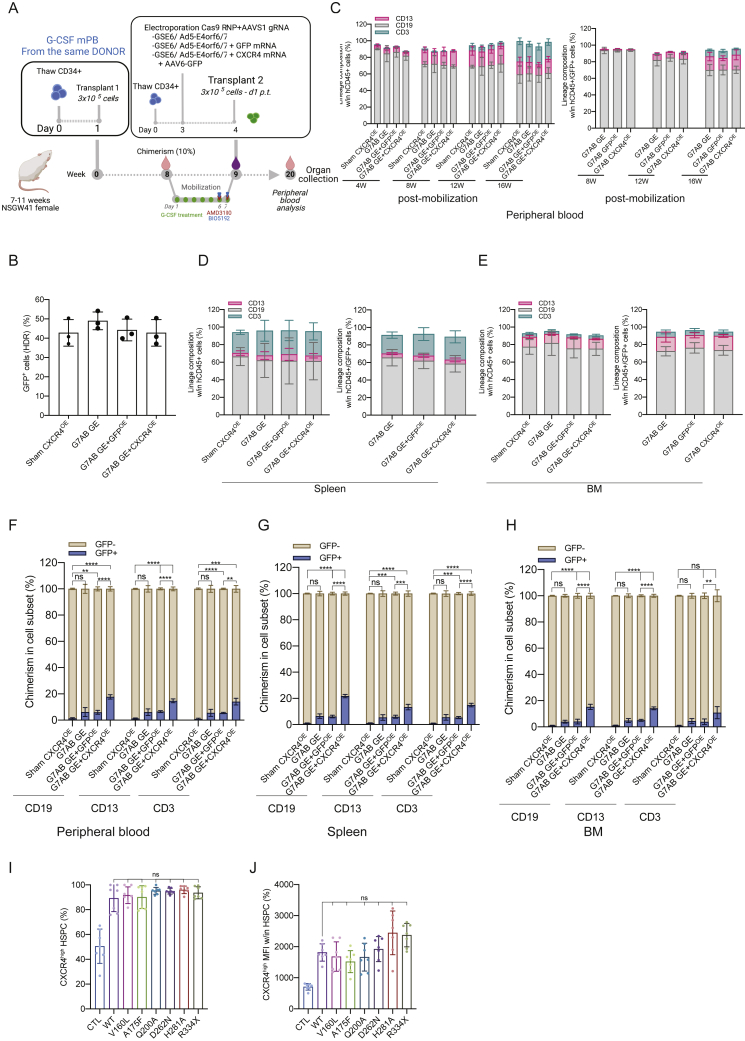
Figure S6.
M-HSCT confers a significant advantage to gene-edited cells when paired with an engraftment enhancer, related to Figure 5
(A) Schematic illustration of competitive transplantation, after G7AB mobilization, between human resident cells (initially transplanted with 3 × 105 CD34+ G-CSF mPB cells, counted on d1 p.t.) and newly transplanted CD34+ G-CSF mPB gene-edited cells (an outgrowth of 3 × 105 CD34+ cells, counted on d1 p.t., and transplanted on d4 p.t.) in NSGW41 mice.
(B) HDR efficiency in edited cells (GFP+) on the bulk CD34+ population, assessed in vitro, 15 days post electroporation.
(C) The reconstitution of myeloid and lymphoid lineages over time of human CD45+ (left panel) and CD45+/GFP+ cells (right panel) in PB after M-HSCT with CD34+ cells gene edited as in main Figure 5A, in NSGW41 mice. The comparison of lineages between human CD45+ and CD45+/GFP+ cells was performed at the last time point by a mixed-effects model (REML), followed by a post hoc analysis with Dunnett’s test (within groups).
(D and E) Myeloid and lymphoid lineage composition of human CD45+ (left panel) and CD45+/GFP+ cells (right panel) in the spleen (D) and BM (E) after M-HSCT with CD34+ cells gene edited as in main Figure 5A, in NSGW41 mice. The comparison of lineages between human CD45+ and CD45+/GFP+ cells was performed by a mixed-effect model (REML), followed by a post hoc analysis with Dunnett’s test (within groups).
(F–H) Chimerism of GFP+ cells was observed within CD19+ B cells, CD13+ myeloid cells, and CD3+ T cells at the end of the experiment in the PB (F), spleen (G), and BM (H) after M-HSCT with CD34+ cells gene edited as in main Figure 5A, in NSGW41 mice. A mixed-effect model (REML), followed by a post hoc analysis with Tukey’s test (between groups) or by post hoc analysis with Dunnett’s test (within groups).
(I) The percentage of CXCR4high cells in HSPCs (CD34+CD133+CD90+), electroporated with CXCR4 variants mRNA. A Kruskal-Wallis test, followed by a post hoc analysis with Dunn’s test.
(J) CXCR4high MFI in HSPCs (CD34+CD133+CD90+), electroporated with CXCR4 variants mRNA. A Kruskal-Wallis test, followed by a post hoc analysis with Dunn’s test.
The results are mean ± SEM, with n ≧ 8 for data collected in vivo on NSGW41 mice, with n ≧ 5 for the data collected in vivo on NSG mice and n ≧ 5 for data collected in vitro (3 different donors). In all the analyses, p less than 0.05 were considered significant (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. “ns” means non-significance).