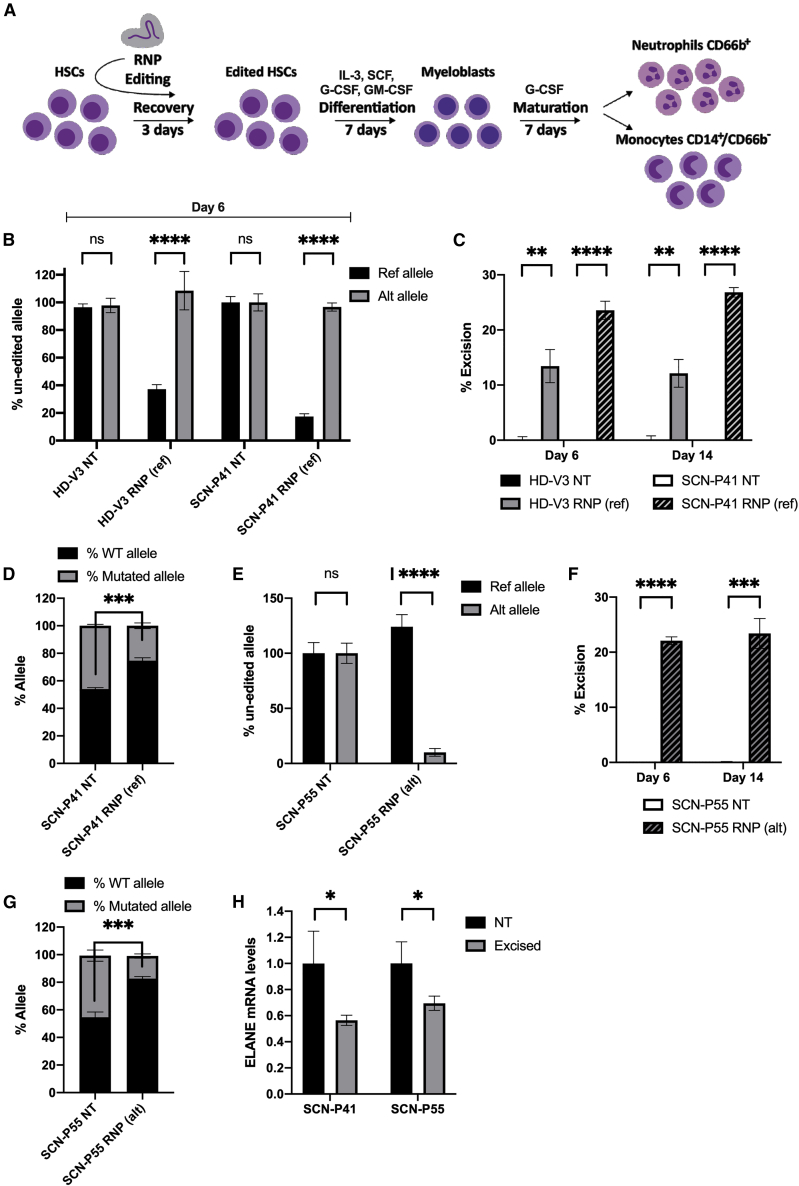
Figure 3.
Allele specificity and excision efficiency of OMNI A1 V10 nuclease compositions
(A) Scheme depicting experimental workflow. HSCs from healthy donors and SCN patients were electroporated with RNPs or left non-treated followed by 3 days of recovery in CD34+ expansion medium. Cells were then subjected to differentiation by culturing for 7 days with IL-3, SCF, GM-CSF, and G-CSF for proliferation and myeloid progenitor differentiation, followed by a 7-day culture in G-CSF for neutrophil differentiation. (B) Bar graph representing percentages of un-edited reference (black) and alternative (gray) alleles at day 6 of differentiation in HSCs taken from either a healthy donor (HD-V3) or an SCN patient (SCN-P41) and treated with RNP(ref) composition or left non-treated (NT), as measured by ddPCR (n = 3 groups of cells from HD-V3 healthy/SCN-P41 patient donors). Statistical significance is indicated by ∗∗∗∗p < 0.0001; ns, not statistically significant. (C) Bar graph representing percentages of excision at days 6 and 14 of differentiation in HSCs taken from either a healthy donor (HD-V3) (non-treated [NT, black] or RNP(ref)-treated [gray]) or an SCN patient (SCN-P41) (NT [white] or RNP(ref)-treated [hatched]), as measured by ddPCR (n = 3 groups of cells from HD-V3 healthy/SCN-P41 patient donors). Statistical significance is indicated by ∗∗p < 0.01, ∗∗∗∗p < 0.0001. (D) Bar graph representing percentages of wild-type (black) and mutated (gray) alleles in cDNA taken from SCN-P41 patient HSCs that were either RNP(ref)-treated or left NT, as measured by NGS targeting the mutation site (n = 3 groups of cells from patient SCN-P41). Statistical significance is indicated by ∗∗∗p < 0.001. (E) Bar graph representing percentages of unedited reference (black) and alternative (gray) alleles at day 6 of differentiation in HSCs taken from an SCN patient (SCN-P55) treated with RNP(alt) composition or left NT, as measured by ddPCR (n = 3 groups of cells from patient donor SCN-P55). Statistical significance is indicated by ∗∗∗∗p < 0.0001; ns, not statistically significant. (F) Bar graph representing percentages of excision at days 6 and 14 of differentiation in HSCs taken from an SCN patient (SCN-P55): NT (white) or RNP(alt)-treated (hatched), as measured by ddPCR (n = 3 groups of cells from patient donor SCN-P55). Statistical significance is indicated by ∗∗∗p < 0.001,∗∗∗∗p < 0.0001. (G) Bar graph representing percentages of wild-type (black) and mutated (gray) alleles in cDNA taken from patient SCN-P55 HSCs that were either RNP(alt)-treated or left NT, as measured by NGS targeting the mutation site (n = 3 groups of cells from patient SCN-P55). Statistical significance is indicated by ∗∗∗p < 0.001. (H) Bar graph representing ELANE mRNA levels in day 6 differentiated HSCs of patients SCN-P41 and SCN-P55 that were either NT (black) or RNP(ref)/RNP(alt)-treated, respectively (excised, gray). Data are presented relative to the NT group (n = 3 groups of cells from patients SCN-P41 and SCN-P55). Statistical significance is indicated by ∗p < 0.05. Bars represent mean values with standard deviation.