Figure 4.
RNP(ref)-facilitated editing boosts neutrophil differentiation and maturation in vitro
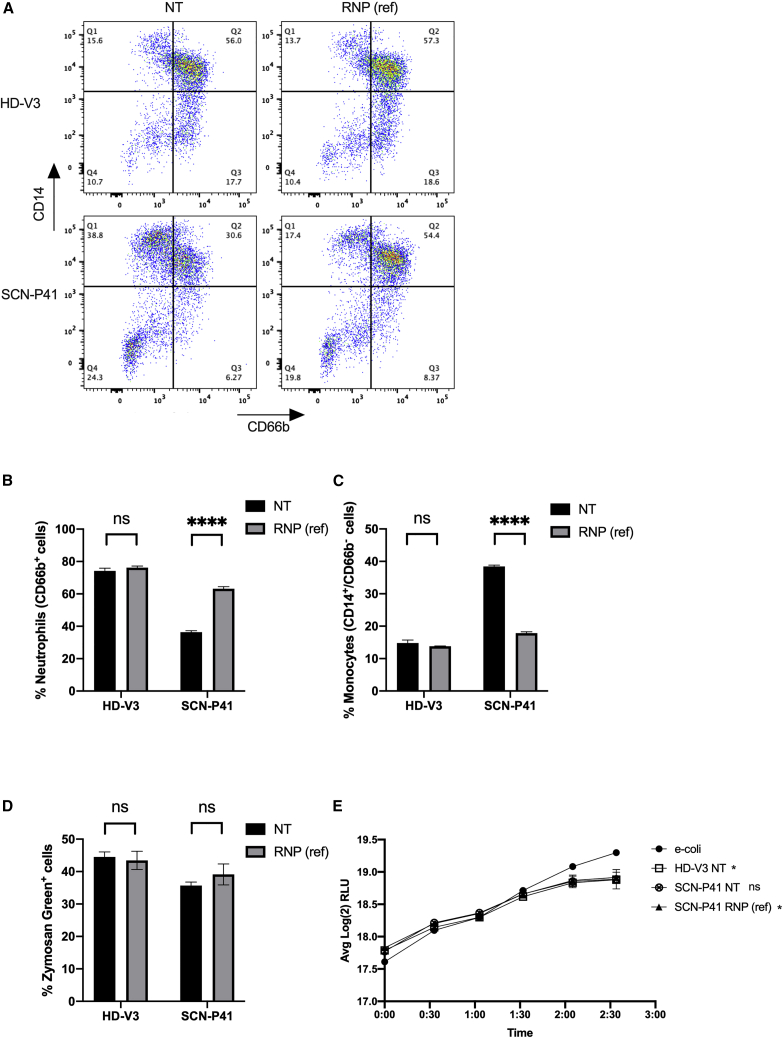
(A) Representative fluorescence-activated cell sorting (FACS) plots of non-treated (NT, left) and RNP(ref)-treated (right) healthy donor (HD-V3, top) and patient SCN-P41 (bottom ) differentiated HSCs, analyzed for neutrophilic (CD66b+) and monocytic (CD14+/CD66b−) subsets. (B) Quantitative analysis of respective FACS data for percentages of neutrophils (CD66b+ cells) in healthy (HD-V3) and SCN patient (SCN-P41) differentiated HSCs that were NT (black) or treated with RNP(ref) (gray) (n = 3 groups of cells from HD-V3 healthy/SCN-P41 patient donors). Statistical significance is indicated by ∗∗∗∗p < 0.0001; ns, not statistically significant. (C) Quantitative analysis of respective FACS data for percentages of monocytes (CD14+/CD66b− cells) in healthy (HD-V3) and SCN patient (SCN-P41) differentiated HSCs that were NT (black) or treated with RNP(ref) (gray) (n = 3 groups of cells from HD-V3 healthy/SCN-P41 patient donors). Statistical significance is indicated by ∗∗∗∗p < 0.0001; ns, not statistically significant. (D) Quantification of percentages of zymosan green uptake by healthy (HD-V3) and SCN patient (SCN-P41) differentiated HSCs that were NT (black) or treated with RNP(ref) (gray). ns, not statistically significant. (E) Graph depicts real-time change in light emission, in relative light units (RLUs), from 200,000 luciferase-expressing bacterial cells incubated with differentiated neutrophils from healthy NT (HD-V3; square), patient NT (SCN-P41; crossed circle), or patient RNP(ref)-treated (SCN-P41, RNP(ref); triangle) HSCs compared with bacterial cells-only control (e-coli; circle). Statistical significance for each of the groups versus the E. coli control at the last time point presented, when RLU levels reached plateau, is indicated by ∗p < 0.05; ∗∗p < 0.01; ns, not statistically significant. Bars represent mean values with standard deviation.