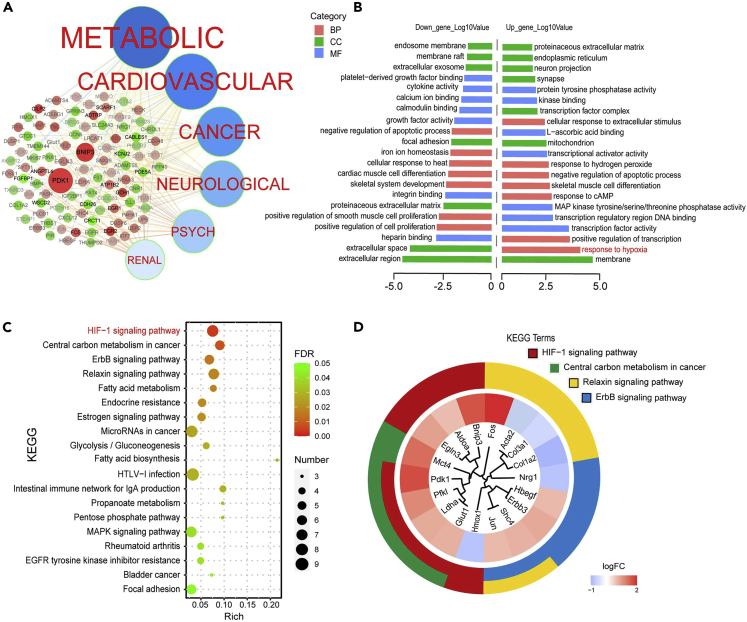
Figure 3.
Transcriptome profiles of HT22 cells treated with myriocin
HT22 cells treated with myriocin (0.5 μM) for 36 h were analyzed for differential expression genes (DEGs, n = 3, p < 0.05).
(A) Disease and DEGs correlation network. The size of the knots represents the p value of DEGs, and the red and green color represent the up- and downregulated genes, respectively.
(B) Gene Ontology (GO) annotation for up- and downregulated DEGs (BP, biological process, CC, cell component, MF, molecular function).
(C) KEGG pathway enrichment analysis of up- and downregulated DEGs. Numbers of enriched genes, enrichment scores, and FDR are presented.
(D) Circular dendrogram of the top four KEGG clusters.