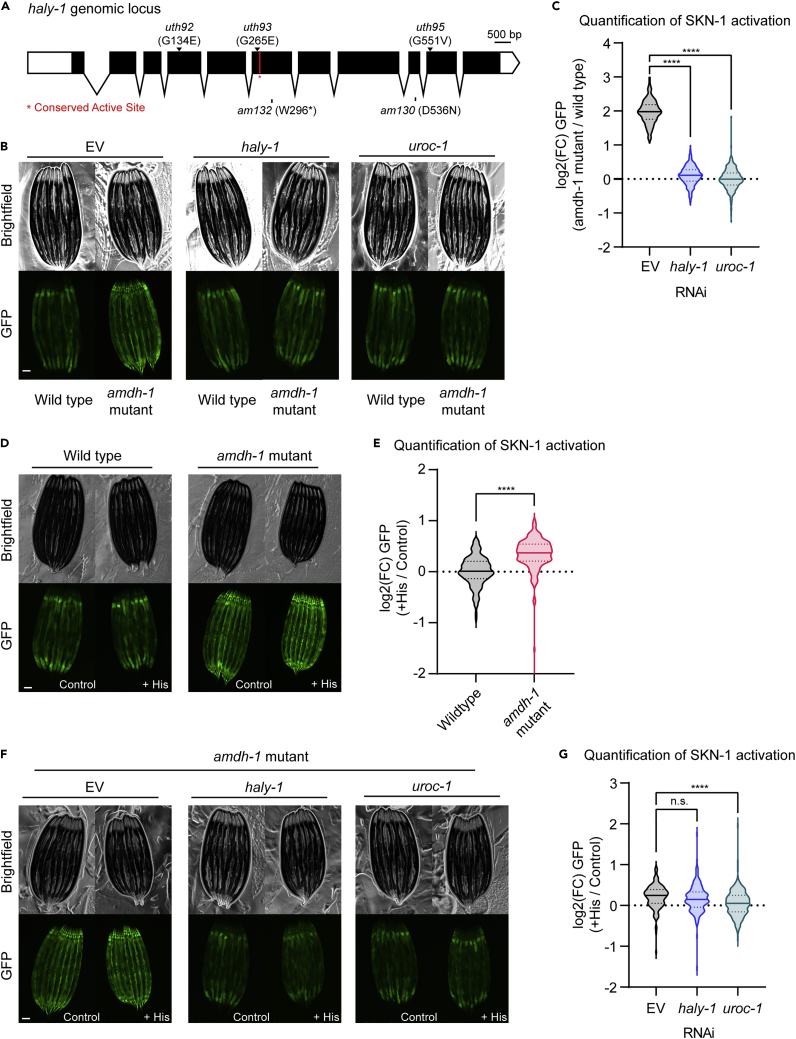
Figure 3.
Activation of SKN-1 requires upstream histidine catabolism enzymes and likely proceeds through the buildup of a metabolic intermediate
(A) Schematic of haly-1 genomic locus labeling novel alleles from EMS screen (top, arrows). A conserved active site is labeled in red and two existing alleles are labeled (bottom). Scale bar, 500 bp.
(B) Fluorescent images of SKN-1 reporter animals in a wildtype or amdh-1(uth29) mutant background fed RNAi targeting haly-1 and uroc-1. Scale bar, 100 μm.
(C) Quantification of SKN-1 activation (amdh-1 mutant normalized to median of wild type) from (B), Data shown are representative of n = 3 biological replicates with n > 250 animals per condition for each replicate. ∗∗∗∗ = p < 0.0001 using a one-way ANOVA.
(D) Fluorescent images of SKN-1 reporter animals in a wildtype or amdh-1(uth29) mutant background with (+His) or without (control) 10mM histidine added to the media. Scale bar, 100 μm.
(E) Quantification of SKN-1 activation (+His normalized to median of control) from (D), Data shown are representative of n = 3 biological replicates with n > 141 animals per condition for each replicate. ∗∗∗∗ = p < 0.0001 Mann-whitney U test.
(F) Fluorescent images of SKN-1 reporter animals in an amdh-1(uth29) mutant background fed RNAi with or without 10mM histidine added to the media. Scale bar, 100 μm.
(G) Quantification of SKN-1 activation (+His normalized to median of control) from (F), Data shown are representative of n = 3 biological replicates with n > 193 animals per condition for each replicate. ∗∗∗∗ = p < 0.0001 one-way ANOVA.