Figure 7.
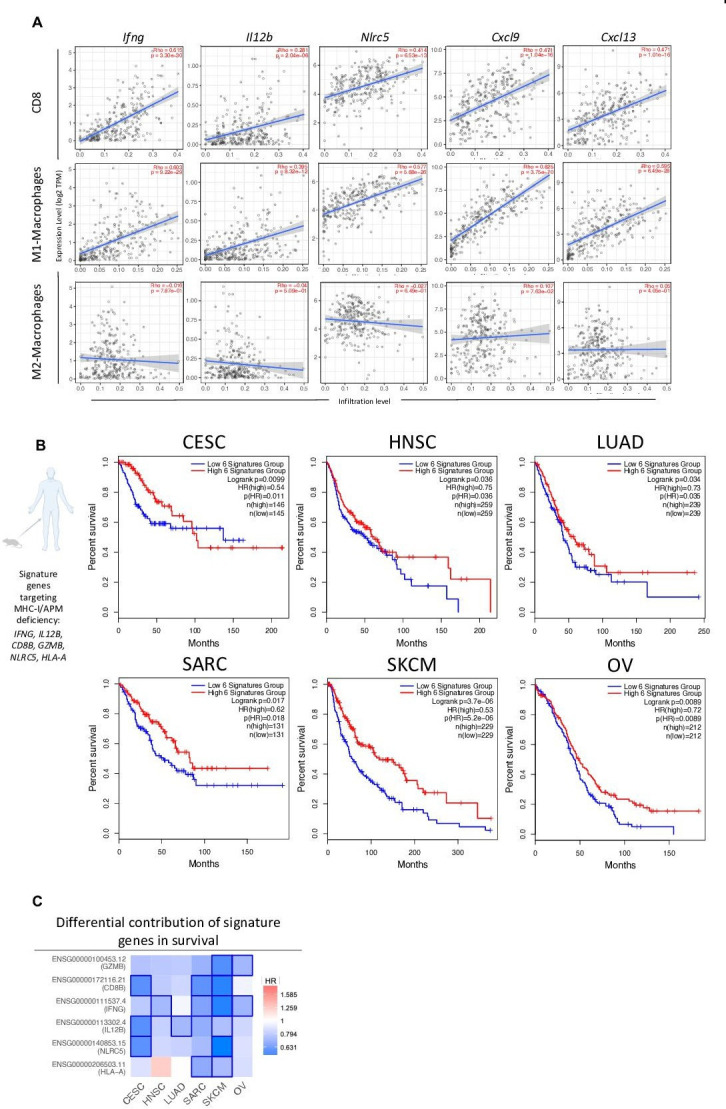
Combination therapy-associated signature shows translational relevance in HPV+ and HPVneg malignancies. TIMER (A) and GEPIA (B–C) TCGA data analysis platforms were used for producing immune correlations and Kaplan-Meier survival curves across multiple cancers. (A) Dot plots of two-tailed Spearman’s correlation of CD8+ T cell, M1- and M2-macrophage tumor infiltration to genes (IFNG, IL12B, NLRC5, CXCL9, CXCL13) encoding combination therapy-induced key immune markers from TCGA CESC dataset (n=291 patients). Spearman’s r >0 indicates positive correlation. Spearman’s r <0 indicates negative correlation. Line represents best-fitting regression line with 95% CIs (gray shading). (B) Correlation between clinical overall survival for designated cancers with key immune signature (IFNG, IL12B, CD8B, GZMB, NLRC5, and HLA-A) observed in murine models of MHC deficiency. (C) HR of key immune signature genes to survival across malignancies. Kaplan-Meier curves were generated with a median high/low 50% cut-off. Mouse and human icons created with BioRender.com. CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CI, confidence interval; GEPIA, Gene Expression Profiling Interactive Analysis; HNSC, head and neck squamous cell carcinoma; HPV, human papilloma virus; HR, hazard ratio; LUAD, lung adenocarcinoma; MHC, major histocompatibility complex; SARC, sarcoma; SKCM, skin cutaneous melanoma; TCGA, The Cancer Genome Atlas; TIMER, Tumor Immune Estimation Resource; OV, ovarian serous cystadenocarcinoma.
