Abstract
Human acanthocephaliasis is a rare parasitic zoonosis mainly caused by acanthocephalans belonging to the genera Acanthocephalus, Bolbosoma, Corynosoma, Macracanthorhynchus, and Moniliformis. In the present paper, the juveniles of Bolbosoma nipponicum Yamaguti, 1939 collected from the northern fur seal Callorhinus ursinus (Linnaeus) (Mammalia: Carnivora) in Alaska, USA were precisely identified based on morphological characters and genetic data. Their detailed morphology was studied using light and, for the first time, scanning electron microscopy. The molecular characterization of the nuclear genes [small ribosomal subunit (18S) and large ribosomal subunit (28S)] and the mitochondrial cytochrome c oxidase subunit 1 (cox1) sequence data of B. nipponicum are provided for the first time. Moreover, in order to clarify the phylogenetic relationships of the genus Bolbosoma and the other genera in the family Polymorphidae, phylogenetic analyses were performed integrating different nuclear (18S + ITS+28S) and mitochondrial (cox1) sequence data using maximum likelihood (ML) and Bayesian inference (BI). The phylogenetic results showed that Bolbosoma has a sister relationship with Corynosoma, and also revealed that Southwellina is sister to Ibirhynchus + Hexaglandula. Our molecular phylogeny also indicated a possible host-switch pattern during the evolution of the polymorphid acanthocephalans. The ancestors of polymorphid acanthocephalans seem to have originally parasitized fish-eating waterfowl in continental habitats, then extended to fish-eating marine birds in brackish water and marine habitats, and finally, opportunistically infected the marine mammals.
Keywords: Zoonotic parasite, Acanthocephala, Human acanthocephaliasis, Molecular identification, Phylogeny
Graphical abstract
Highlights
-
•
Detailed morphology of the juveniles of B. nipponicum was described for the first time.
-
•
Molecular characterization of the 18S, 28S and cox1 genes of B. nipponicum was provided for the first time.
-
•
Molecular phylogenetic analyses showed that Bolbosoma has a sister relationship with Corynosoma.
1. Introduction
The current species identification of acanthocephalans is still mainly based on morphological methods. Recent studies have indicated that it is useful and practical to utilize some nuclear and mitochondrial DNA sequences [i.e., large ribosomal DNA (28S), internal transcribed spacer (ITS) and cytochrome oxidase subunit 1 (cox1)] as genetic markers for the accurate identification of acanthocephalans, especially for the delimitation of intraspecific phenotypic variation, identification of cystacanths/juveniles and discovery of cryptic species (Alcántar-Escalera et al., 2013; Kang and Li, 2018; Li et al., 2019; Lisitsyna et al., 2019; Steinauer et al., 2007; Wayland et al., 2015; Zittel et al., 2018). However, most of the currently recognized acanthocephalan species were defined only under the traditional morphospecies concept.
The genus Bolbosoma Porta, 1908 (Palaeacanthocephala: Polymorphida) is a small group of acanthocephalans, with adults mainly parasitic in marine mammals, i.e., seals, whales and dolphins (Amin and Margolis, 1998; Baylis, 1929; Delyamure, 1955; Santoro et al., 2021; Skrjabin, 1972; Wang, 1980; Yamaguti, 1939). Amin (2013) listed 12 nominal species, namely B. australis Skrjabin, 1972, B. balaenae (Gmelin, 1790), B. brevicolle (Malm, 1867), B. caenoforme (Heitz, 1920), B. capitatum (von Linstow, 1880), B. hamiltoni Baylis, 1929, B. heteracanthe (Heitz, 1920), B. nipponicum Yamaguti, 1939, B. scomberomori Wang, 1980, B. tuberculata Skriabin, 1970, B. turbinella (Diesing, 1851) and B. vasculosum (Rudolphi, 1819). Some species of Bolbosoma are recognized as parasites that are frequently associated with human acanthocephaliasis (Beaver et al., 1983; Hino et al., 2002; Ishikura et al., 1996; Isoda et al., 2006; Kaito et al., 2019; Mori et al., 1998; Tada et al., 1983). To date, at least nine cases of human acanthocephaliasis caused by Bolbosoma spp. have been reported in Japan (Arizono et al., 2012; Kaito et al., 2019; Santoro et al., 2021). Humans become infected by the accidental ingestion of raw or undercooked marine fishes/squids contaminated by cystacanths of Bolbosoma (Kaito et al., 2019; Tada et al., 1983).
The adults of B. nipponicum were originally described from the common minke whale Balaenoptera acutorostrata Lacépède (Mammalia: Cetacea) in the North Pacific Ocean (Yamaguti, 1939). Later, this species was reported in various marine mammals, including the northern fur seal Callorhinus ursinus (Linnaeus) (Mammalia: Carnivora) (Delyamure 1955; Kuzmina et al., 2012, 2021). However, the detailed morphology of the juveniles of B. nipponicum has never been described. Furthermore, the current genetic data base for this species is still scanty, with only the partial ITS rDNA available in the GenBank now (Arizono et al., 2012).
In the present study, the juveniles of B. nipponicum were described using light and, for the first time, scanning electron microscopy, based on specimens collected from the northern fur seal C. ursinus in St. Paul Island, Alaska, USA. The molecular characterization of the small ribosomal DNA (18S), the large ribosomal DNA (28S) and the mitochondrial cytochrome c oxidase subunit 1 (cox1) sequence data of B. nipponicum are provided for the first time. Moreover, in order to clarify the phylogenetic relationships of the genus Bolbosoma and the other genera in the family Polymorphidae Meyer, 1931, phylogenetic analyses were performed integrating different nuclear (18S + ITS + 28S) and mitochondrial (cox1) genetic markers using maximum likelihood (ML) and Bayesian inference (BI).
2. Materials and methods
2.1. Morphological study
Acanthocephalans were collected from the intestine of northern fur seal C. ursinus in St. Paul Island, Alaska, USA in 2014 (see Kuzmina et al., 2021 for details); fresh helminths were washed in tap water, then fixed and preserved in 70% ethanol. For light microscopical studies, three juvenile acanthocephalans were placed in temporary mounts and cleared in glycerin. Photomicrographs were recorded using a Nikon® digital camera coupled to a Nikon® optical microscopy (Nikon ECLIPSE Ni–U, Nikon Corporation, Tokyo, Japan). For scanning electron microscopy (SEM), the anterior part of one juvenile specimen was post-fixed in 1% OsO4, dehydrated via an ethanol series and acetone, and then critical point dried. The specimen was coated with gold at about 20 nm and examined using a Hitachi S-4800 scanning electron microscope at an accelerating voltage of 20 kV. Measurements (the range, followed by the mean in parentheses) are given in micrometres unless otherwise stated.
2.2. Molecular procedures
The mid-body of one male and one female juvenile specimens were used for molecular analysis (the anterior and posterior ends of body deposited for morphological study). Genomic DNA from each sample was extracted using a Column Genomic DNA Isolation Kit (Shanghai Sangon, China) according to the manufacturer's instructions. The partial 18S region was amplified in one fragment by polymerase chain reaction (PCR) using forward primer (5′-AGATTAAGCCATGCATGCGTAAG-3′) and the reverse primer (5′-TGATCCTTCTGCAGGTTCACCTAC-3′) (Garey et al., 1996). The partial 28S region was amplified in 4 overlapping fragments (amplicons 1–4) by PCR using the following primers (García-Varela and Nadler, 2005), including amplicon 1: forward 5′-CAAGTACCGTGAGGGAAAGTTGC-3′ and reverse 5′-CAGCTATCCTGAGGGAAAC-3′; amplicon 2: forward 5′-ACCCGAAAGATGGTGAACTATG-3′ and reverse 5′-CTTCTCCAAC(T/G)TCAGTCTTCAA-3′; amplicon 3: forward 5′-CTAAGGAGTGTGTAACAACTCACC-3′ and reverse 5′-AATGACGAGGCATTTGGCTACCTT-3′; amplicon 4: forward 5′-GATCCGTAACTTCGGGAAAAGGAT-3′ and reverse 5′-CTTCGCAATGATAGGAAGAGCC-3′. The partial ITS region was amplified by PCR using the forward primer ITS-F (5′-GTC GTA ACA AGG TTT CCG TA-3′) and the reverse primer ITS-R (5′-TAT GCT TAA ATT CAG CGG GT-3′) (Král’ová-Hromadová et al., 2003). The partial cox1 region was amplified by PCR using the forward primer COI–F (5′-GGT CAA CAA ATC ATA AAG ATA TTG G-3′) and the reverse primer COI-R (5′-TAA ACT TCA GGG TGA CCA AAA AAT CA-3′) (Gómez et al., 2002). Cycling conditions were as previously described by (Li et al., 2019). PCR products were checked on GoldView-stained 1.5% agarose gels and purified with Column PCR Product Purification Kit (Shanghai Sangon, China). Sequencing of each sample was carried out for both strands. Sequences were aligned using ClustalW2. The DNA sequences obtained herein were compared (using the algorithm BLASTn) with those available in the National Center for Biotechnology Information (NCBI) database (http://www.ncbi.nlm.nih.gov). The 18S, ITS, 28S and cox1 sequences of B. nipponicum were deposited in the GenBank database (http://www.ncbi.nlm.nih.gov) (accession numbers: 18S: ON358429, ON361558; ITS: ON361563, ON361566; 28S: ON359851, ON359856; cox1: ON359908, ON359909).
2.3. Phylogenetic analyses
Phylogenetic trees were constructed based on the 18S + ITS+28S + cox1 sequence data using maximum likelihood (ML) inference with IQTREE (Nguyen et al., 2015) and Bayesian inference (BI) with Mrbayes 3.2.7 (Ronquist et al., 2012), respectively. Centrorhynchus clitorideus (Palaeacanthocephala: Polymorphida: Centrorhynchidae) was chosen as the out-group according to the previous studies (García-Varela et al., 2013). The in-group includes 25 polymorphid species representing 10 genera with sequences available on GenBank. The detailed information on acanthocephalan species included in the present phylogenetic analyses was provided in Table 1. The nucleotide sequences of each gene were aligned in batches using MAFFT v7.313 under iterative refinement method of E–INS–I (Katoh and Standley, 2013), poorly aligned regions were excluded using BMGE v1.12 (h = 0.4) (Criscuolo and Gribaldo, 2010). Further, partially ambiguous bases were manually inspected and removed. PhyloSuite v1.2.2 (Zhang et al., 2020) was then used to concatenate these alignments into a single alignment and generate phylip and nexus format files for the phylogenetic analyses.
Table 1.
Detailed information of representatives of the family Polymorphidae used for phylogenetic analyses.
The maximum likelihood inference was conducted in IQTREE v2.1.2 (Minh et al., 2020). Substitution models were compared and selected according to the Bayesian Information Criterion by using ModelFinder (Kalyaanamoorthy et al., 2017). The TVM + F + R3 model was identified as the optimal nucleotide substitution model for 18S + 28S + ITS + cox1 sequence data. Reliabilities for maximum likelihood inference were tested using 1000 bootstrap replications and Bayesian Information Criterion analysis was run for 5 × 106 MCMC generations and sampling a tree with every 1000 generations. The first 25% trees were treated as “burn-in”. The phylogenetic trees were visualized in iTOL v6.1.1 (Letunic and Bork, 2021). In the ML tree, the bootstrap values ≥ 85 were considered to constitute strong nodal support, whereas BS values ≥ 50 and <85 were considered to constitute moderate nodal support. In the BI tree, the Bayesian posterior probabilities values ≥ 0.90 were considered to constitute strong nodal support, whereas Bayesian posterior probabilities values ≥ 0.70 and < 0.90 were considered to constitute moderate nodal support. The bootstrap values ≥ 50 and Bayesian posterior probabilities values ≥ 0.70 were shown in the phylogenetic trees.
3. Results
3.1. Morphology of juveniles of Bolbosoma nipponicumYamaguti, 1939 (Fig. 1, Fig. 2, Fig. 3)
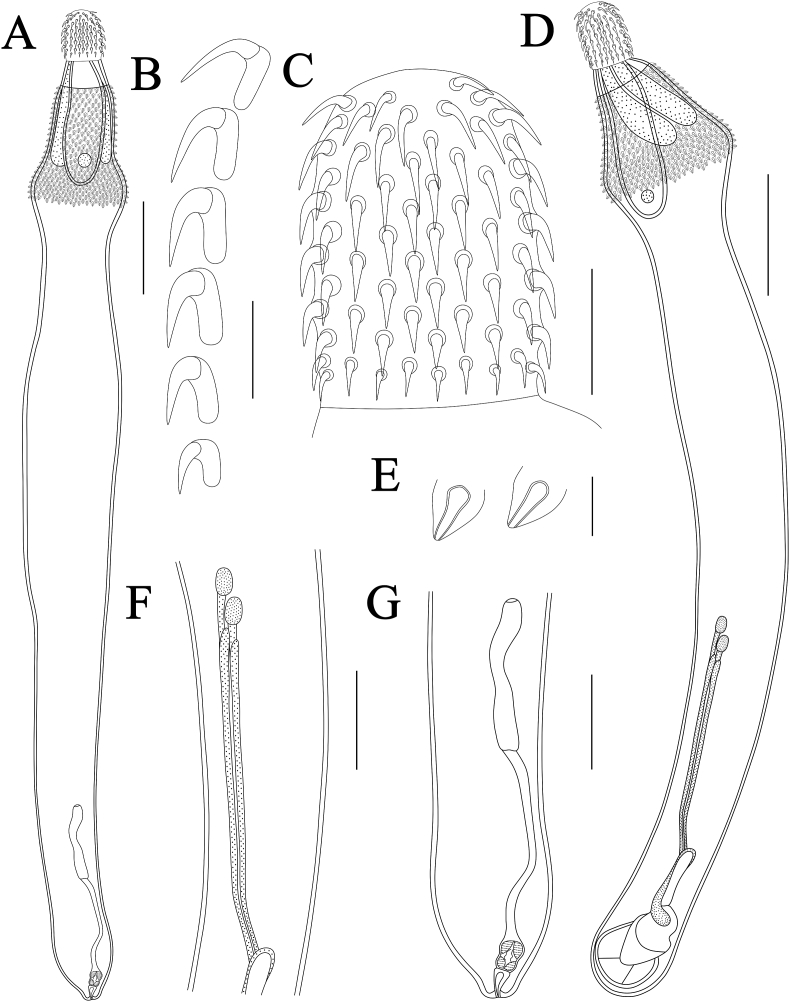
Fig. 1.
Bolbosoma nipponicum collected from Callorhinus ursinus (Linnaeus) (Carnivora: Otariidae) in St. Paul Island, Alaska. A: female; B: hooks; C: proboscis; D: male; E: trunk spines; F: testes and cement-glands; G: poster part of female. Scale bars: A, D = 1000 μm; B = 100 μm; C = 200 μm; E = 50 μm; F, G = 500 μm.
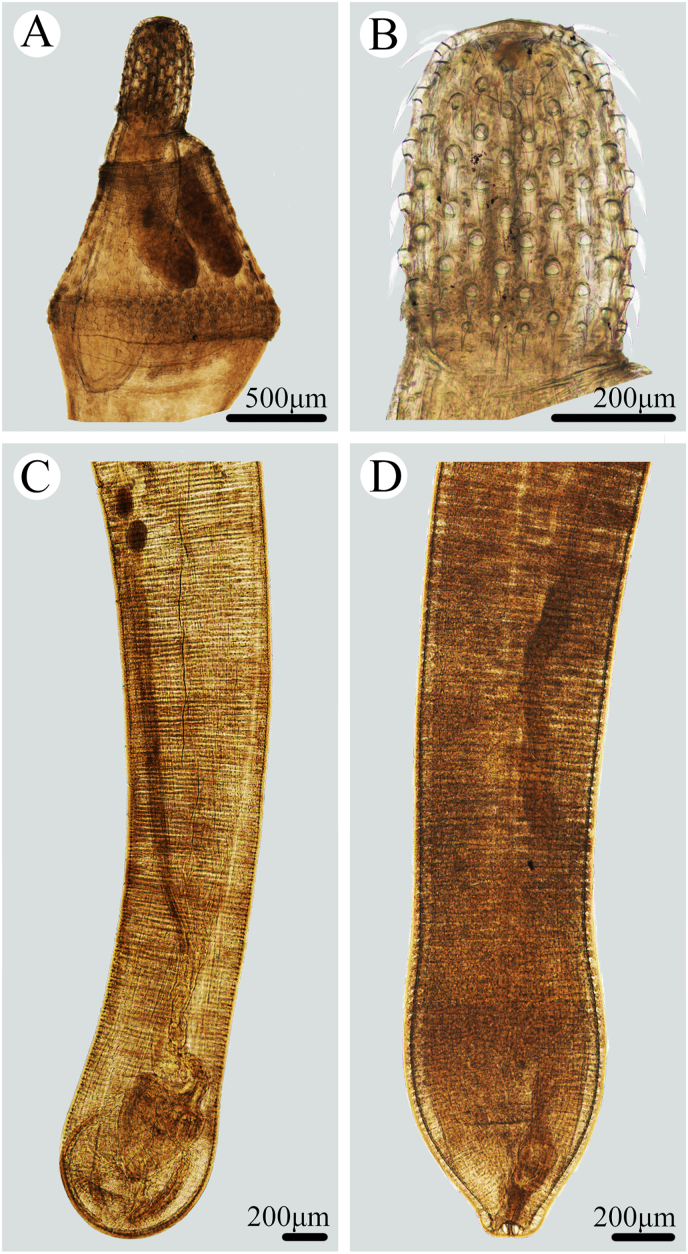
Fig. 2.
Photomicrographs of Bolbosoma nipponicum collected from Callorhinus ursinus (Linnaeus) (Carnivora: Otariidae) in St. Paul Island, Alaska. A: anterior part of male; B: proboscis; C: posterior part of male; D: posterior part of female.
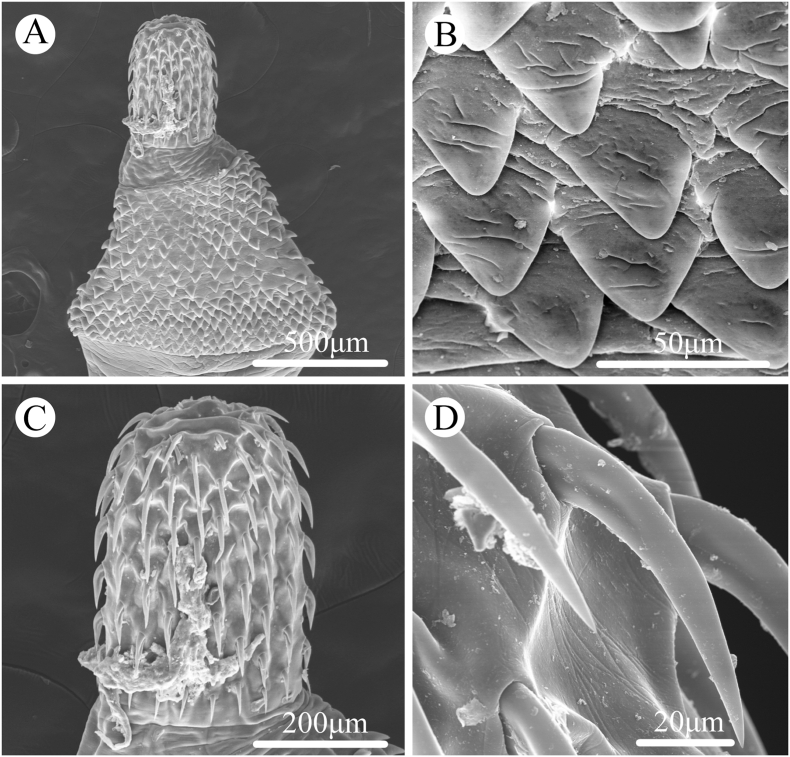
Fig. 3.
Scanning electron micrographs of Bolbosoma nipponicum collected from Callorhinus ursinus (Linnaeus) (Carnivora: Otariidae) in St. Paul Island, Alaska. A: anterior part of male; B: trunk spines; C: proboscis; D: hooks.
General. Trunk medium, more or less cylindrical, elongate, with bulbous enlargement in anterior part (Fig. 1, Fig. 2, Fig. 3A). Trunk spines scaly, extending from base of neck to just posterior of maximum enlargement (Fig. 1, Fig. 2, Fig. 3A, B). Proboscis cylindrical, with 20–22 longitudinal rows of 6–7 hooks per row (Fig. 1, Fig. 2, Fig. 3A, C, D). Apical proboscis hooks slender, middle hooks robust, basal hooks smallest, all hooks with simple posteriorly directed roots (Fig. 1, Fig. 2, Fig. 3A, C, D). Proboscis receptacle double-walled, cerebral ganglion at base of proboscis receptacle (Fig. 1, Fig. 2A). Neck trapezoid (Fig. 1, Fig. 2, Fig. 3A). Lemnisci subequal, stubby, distinctly shorter than proboscis receptacle (Fig. 1, Fig. 2A).
Male [Based on 2 juvenile male specimens] (Table 2). Trunk 7.48–8.35 (7.91) mm long, maximum width 1.08–1.23 (1.15) mm. Proboscis 525–584 (554) long, 376–416 (396) wide. Size of proboscis hooks from anterior: 88–93 × 18–19 (90 × 18); 93–98 × 18–23 (95 × 20); 93–95 × 18–23 (94 × 20); 93–95 × 23–28 (94 × 25); 75–80 × 20–23 (78 × 21); 53–55 × 13–15 (54 × 14). Neck 158–188 (173) long, 485–525 (505) wide. Proboscis receptacle 1.22–1.33 (1.28) mm long, 416–475 (446) wide. Longer lemniscus 446–851 (648) long, 158–238 (198) wide. Shorter lemniscus 396–822 (609) long, 119–178 (149) wide. Testes two, very small, oval; nearly equal in size, 141–169 (154) long, 80–127 (105) wide (Fig. 1, Fig. 2C). Cement-glands two pairs, slender, tubular, 1.68–2.18 (1.93) mm long, 30–40 (35) wide (Fig. 1, Fig. 2C). Copulatory bursa not everted, 802–822 (812) long, 525–545 (535) wide. Gonopore terminal.
Table 2.
Comparative morphometric data for Bolbosoma nipponicum (all measurements are in millimetres).
| Hosts |
Callorhinus ursinus |
Balaenoptera rostrata |
Balaenoptera borealis |
|||
|---|---|---|---|---|---|---|
| Localities |
Alaska, USA |
North Pacific Ocean |
North Pacific Ocean |
|||
| Sources |
Present study |
Yamaguti (1939) |
Fukui and Morisita (1939) |
|||
| Characteristics/sex | Immature male | Immature female | Mature male | Mature female | Mature male | Mature female |
| Trunk length | 7.48–8.35 | 9.93 | up to 45.0 | up to 60.0 | 20.0–28.0 | 25.0–33.0 |
| Trunk width | 1.08–1.23 | 1.13 | 1.20–4.00 | 1.20–4.00 | – | – |
| Proboscis length | 0.52–0.58 | 0.59 | 0.40–0.65 | 0.40–0.65 | 0.86 | 0.86 |
| Proboscis width | 0.38–0.42 | 0.45 | 0.30–0.44 | 0.30–0.44 | 0.40 | 0.40 |
| Hook rows | 20–22 | 22 | 17–23 | 17–23 | 19–21 | 19–21 |
| Hooks/per row | 6–7 | 6 | 5–6 | 5–6 | 7–8 | 7–8 |
| Lemnisci length | 0.40–0.85 | 1.12–1.16 | 1.00–2.30 | 1.00–2.30 | – | – |
| Lemnisci width | 0.12–0.24 | 0.19–0.23 | 0.12–0.30 | 0.12–0.30 | – | – |
| Proboscis receptacle length | 1.22–1.33 | 1.35 | 1.10–1.75 | 1.10–1.75 | 1.65 | 1.65 |
| Proboscis receptacle width | 0.42–0.48 | 0.47 | 0.35–0.48 | 0.35–0.48 | 0.55 | 0.55 |
| Neck length | 0.16–0.19 | 0.35qw | 0.50–0.60 | 0.50–0.60 | – | – |
| Neck width | 0.49–0.52 | 0.54 | 0.62–0.80 | 0.62–0.80 | – | – |
| Testis length | 0.14–0.17 | – | 1.00–2.00 | – | – | – |
| Testis width | 0.08–0.13 | – | 0.60–1.05 | – | – | – |
| Cement gland length | 1.68–2.18 | – | 2.70 | – | – | – |
| Size of egg | – | – | – | 0.12–0.19 × 0.03–0.04 | – | 0.15 × 0.03 |
| Uterus length | – | 1.07 | – | 0.23–3.85 | – | – |
Female [Based on 1 juvenile female specimen] (Table 2). Trunk 9.93 mm long, maximum width 1.13 mm. Proboscis 594 long, 446 wide. Size of proboscis hooks from anterior to posterior: 79 × 18; 103 × 21; 101 × 25; 91 × 24; 82 × 21; 59 × 15. Neck 347 long, 545 wide. Proboscis receptacle 1.35 mm long, 465 wide. Longer lemnisci 1.16 mm long, 228 wide. Shorter lemnisci 1.12 mm long, 188 wide. Uterine bell 842 long, 139 wide. Uterus 1.07 mm long, 30 wide, vagina 327 long, 129 wide (Fig. 1, Fig. 2D). Eggs not observed. Gonopore terminal (Fig. 1, Fig. 2D).
Hosts of present material: Callorhinus ursinus (Linnaeus) (Carnivora: Otariidae).
Locality: St. Paul Island, Alaska, USA (57° 09′ N, 170° 13′ W).
Site infection: Intestine.
Voucher specimens: 2 juvenile males, 1 juvenile female (HBNU–A-2022M002L), deposited in the College of Life Sciences, Hebei Normal University, Hebei Province, P. R. China.
3.2. Molecular characterization
3.2.1. Partial 18S region
Two 18S sequences of B. nipponicum obtained herein are both 1182 bp in length, with no nucleotide divergence detected. In the genus Bolbosoma, the 18S sequence data are available in GenBank for B. turbinella (JX442166), B. balaenae (MZ047218–MZ047227, JQ040304–JQ040306, MT233305), B. vasculosum (JX014225), B. caenoforme (KF156879) and Bolbosoma sp. MGV-2012 (JX442167). Pairwise comparison of 18S sequences of B. nipponicum with these five species of Bolbosoma produced 0–0.42% of nucleotide divergence.
3.2.2. Partial 28S region
Two 28S sequences of B. nipponicum obtained herein are both 2755 bp in length, with 0.04% nucleotide divergence detected. In the genus Bolbosoma, the 28S sequence data are available in GenBank for B. turbinella (JX442178), B. balaenae (MZ047231–MZ047239) and Bolbosoma sp. MGV-2012 (JX442179). Pairwise comparison of 28S sequences of B. nipponicum and these three species of Bolbosoma produced 0–1.12% (B. turbinella) of nucleotide divergence.
3.2.3. Partial ITS region
Two ITS sequences of B. nipponicum obtained herein are both 754 bp in length, with 0.13% nucleotide divergence detected. In the genus Bolbosoma, the ITS sequence data are available in GenBank for B. nipponicum (AB706183), B. cf. capitatum (AB706182), B. turbinella (KU314817–KU314819) and Bolbosoma sp. KMC (LC375174) (Table 1). Pairwise comparison of ITS sequences of B. nipponicum obtained herein with B. nipponicum (AB706183) available in GenBank displayed 0–0.13% of nucleotide divergence, and with the other three species of Bolbosoma produced 9.42–11.6% of nucleotide divergence.
3.2.4. Partial cox1 region
Two cox1 sequences of B. nipponicum obtained herein are both 655 bp in length, with 0.31% nucleotide divergence detected. In the genus Bolbosoma, the cox1 sequence data are available in GenBank for B. turbinella (JX442189, KU314821, KU314823), B. balaenae (MZ047272–MZ047281), B. caenoforme (KF156891) Bolbosoma sp. MJA-2016 (KX098556) and Bolbosoma sp. KMC (LC377776) (Table 1). Pairwise comparison of cox1 sequences of B. nipponicum with B. caenoforme (KF156891) showed 0.15% of nucleotide divergence, and with the other four species of Bolbosoma produced 13.9–31.7% of nucleotide divergence.
3.3. Phylogenetic analyses (Fig. 4, Fig. 5)
Fig. 4.
Phylogenetic relationships of representatives of the family Polymorphidae using maximum likelihood method based on the 18S + ITS +28S + cox1 sequence data. Centrorhynchus clitorideus (Polymorphida: Centrorhynchidae) was chosen as outgroup. Bootstrap values > 50 are shown in the phylogenetic tree.
Fig. 5.
Phylogenetic relationships of representatives of the family Polymorphidae using Bayesian inference based on the 18S + ITS +28S + cox1 sequence data. Centrorhynchus clitorideus (Polymorphida: Centrorhynchidae) was chosen as outgroup. Bayesian posterior probabilities values > 0.70 are shown in the phylogenetic tree.
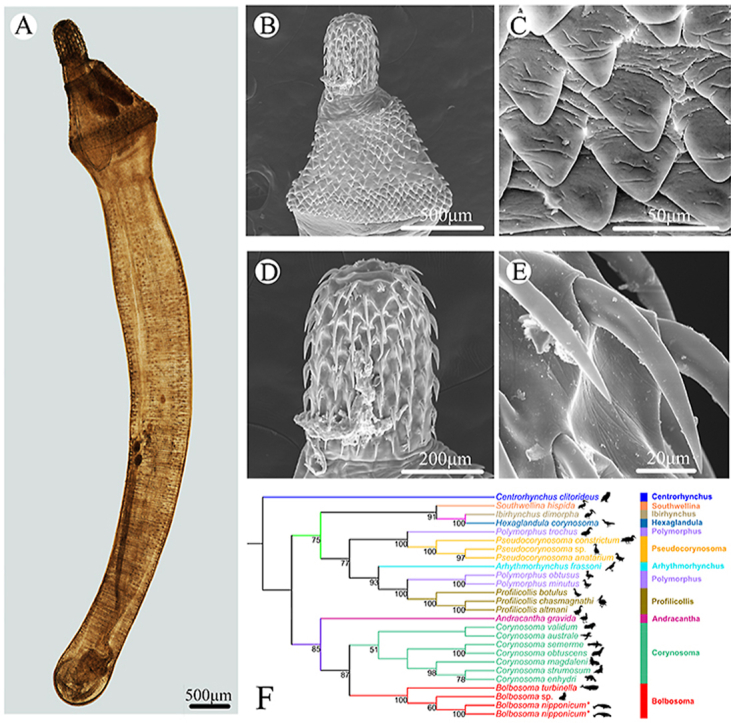
The phylogenetic results based on the 18S + ITS+28S + cox1 sequence data using ML and BI methods, respectively, were very similar (Fig. 4, Fig. 5). In the ML tree (Fig. 4), Southwellina hispida clustered together with the representatives of Ibirhynchus + Hexaglandula with strong support, which formed a sister relationship with all the representatives of Pseudocorynosoma + Arhythmorhynchus + Profilicollis + Polymorphus with moderate support. Species of Bolbosoma clustered together with the representatives of Corynosoma with strong support, which constituted a sister group with Andracantha gravida (Fig. 4). In the BI tree (Fig. 5), Southwellina hispida was at the base of the tree, forming a sister clade to the remaining Polymorphidae with low support value. The representatives of Ibirhynchus + Hexaglandula formed a sister relationship with strong support. The representatives of Pseudocorynosoma + Arhythmorhynchus + Profilicollis + Polymorphus constituted a separated branch with strong support. Species of Bolbosoma also showed sister relationship with the representatives of Corynosoma, both clustered together with A. gravida with strong support.
4. Discussion
Arizono et al. (2012) reported the ITS sequence data of B. nipponicum based on the adult specimen collected from the type host (Balaenoptera acutorostrata) in the type locality (North Pacific Ocean), which enabled us to more accurately identify our present material. Pairwise comparison of ITS sequences of our material and B. nipponicum (AB706183) sequenced by Arizono et al. (2012), showed 0–0.13% of nucleotide divergence, which is accordant with the level of intraspecific genetic variation between different individuals detected herein. Consequently, we considered our material to be conspecific with B. nipponicum.
Although Kuzmina et al. (2012, 2021) found B. nipponicum in the northern fur seal C. ursinus, they did not provide a detailed morphological description of their specimens. Because the present specimens are all juvenile male (testis very small) and female (no eggs found in the uterus), we speculated that the northern fur seal C. ursinus possibly acts as an unsuitable final host for B. nipponicum, or the juvenile worms have only recently infected the northern fur seal and have not had time to mature. The similar situation also occurred in B. vasculosum. Costa et al. (2000) reported the juvenile worms of B. vasculosum from the stranded common dolphin Delphinus delphis in the Atlantic Ocean. In spite of our present specimens being immature, their morphology and morphometrics are more or less identical to the original description of mature adults regarding several features, including the morphology of the neck and trunk, the arrangement of trunk spines, the number and arrangement of the proboscis hooks and the size of proboscis receptacle. However, comparison with Yamaguti's (1939) material, our specimens have distinctly smaller trunk and testis, and shorter trunk spines region and neck (see Table 2 for details). Moreover, the proboscis receptacle is much longer than the lemnisci in the present juvenile specimens (vs the proboscis receptacle distinctly shorter than the lemnisci in the mature specimens), and the maximum enlargement of trunk is distinctly narrower than the mature material of Yamaguti (1939). The shape of proboscis of the juvenile is also slightly different from that of mature specimens. Fukui and Morisita (1939) redescribed B. turbinella based on specimens collected from Balaenoptera borealis Lesson (Cetacea: Balaenopteridae). However, Yamaguti (1939) considered that Fukui and Morisita's (1939) material identified as B. turbinella may be the species B. nipponicum. We also compared the morphology and measurements of these two species (see Table 2 for details) and did not find remarkable differences between Fukui and Morisita's (1939) material and Yamaguti's (1939) specimens, except proboscis of Fukui and Morisita's (1939) material with 7–8 hooks per row (vs proboscis usually with 6 hooks per row in Yamaguiti's specimens).
The molecular characterization of the 18S, 28S and cox1 genes of B. nipponicum are provided for the first time. Our molecular analysis revealed that the level of intraspecific genetic variation in the 18S and 28S sequence data is distinctly lower than that of the interspecific genetic variation in the ITS and cox1 regions. It seems more useful and practical to utilize the ITS and/or cox1 regions than the 18S and 28S sequences as genetic markers for identifying and distinguishing acanthocephalans, especially for the diagnosis of the closely related species. However, we noted that a pairwise comparison of cox1 sequences of our specimens and B. caenoforme (KF156891) provided by Malyarchuk et al. (2014), showed only 0.15% of nucleotide divergence. The voucher specimen of B. caenoforme (KF156891) in Malyarchuk et al. (2014) was collected from the Salvelinus malma (Walbaum) (Salmoniformes: Salmonidae) in the Taui Gulf, which acts as a paratenic host for species of Bolbosoma (Santoro et al., 2021). Although Malyarchuk et al. (2014) claimed their samples to be B. caenoforme, this species identification is almost certainly erroneous. Due to the scarcity of the material of this species, although the numbers of our present specimens are limited, the morphological and genetic data of B. nipponicum presented here will be a valuable reference for future studies on the diagnosis of different developmental stages, population genetics and phylogenetics of this group.
The family Polymorphidae is a large group of acanthocephalans, currently including over 120 species mainly parasitic in marine mammals, fish-eating marine birds and waterfowls (Amin, 2013; Aznar et al., 2006; Delyamure, 1955; Dimitrova and Georgiev, 1994; García-Varela et al., 2011, 2013; Schmidt, 1975). Amin (2013) listed 12 genera in this family, namely Andracantha, Ardeirhynchus, Arhythmorhynchus, Bolbosoma, Corynosoma, Diplospinifer, Filicollis, Ibirhynchus, Polymorphus, Profilicollis, Pseudocorynosoma and Southwellina. Some previous phylogenetic studies supported Polymorphidae to be a monophyletic group (García-Varela and Pérez-Ponce de León, 2008; Martín García-Varela et al., 2011, 2013; Verweyen et al., 2011). Amin (1992, 2013) considered the genus Hexaglandula Petrochenko, 1950 to be a synonym of Polymorphus. However, the recent molecular phylogenetic studies supported that Hexaglandula represents an independent valid genus (García-Varela et al., 2009, 2011, 2013; García-Varela and Pérez-Ponce de León, 2008; Presswell et al., 2017).
Our phylogenetic results using ML and BI methods both showed that the genus Bolbosoma has a sister relationship with Corynosoma with strong support, which agreed well with the recent molecular studies based on 28S + cox1 (Presswell et al., 2017) and 18S + 28S + cox1 sequence data (García-Varela et al., 2013). The close relationship of Bolbosoma and Corynosoma can be easily understood, when we considered they have the same type of definitive hosts. In the family Polymorphidae, only adults of Bolbosoma and Corynosoma can parasitize marine mammals, and some species of Bolbosoma and Corynosoma [i.e., B. capitatum, Bolbosoma sp., C. strumosum (Rudolphi, 1802), C. validum Van Cleave, 1953 and C. villosum Van Cleave, 1953] are important zoonotic pathogens for human acanthocephaliasis (Arizono et al., 2012; Fujita et al., 2016; Kaito et al., 2019). The present phylogenetic results also supported the validity of the genus Hexaglandula, which is sister to Ibirhynchus with strong support in both ML tree and BI tree. The results are accordant with the previous phylogenetic analyses (García-Varela et al., 2013; Presswell et al., 2017). However, our results revealed the genus Southwellina formed a sister clade to the remaining representatives of Polymorphidae with low support value in the BI tree and constituted a sister relationship with Ibirhynchus + Hexaglandula with strong support only in the ML tree, which is different from all the previous phylogenetic studies (García-Varela et al., 2009, 2011, 2013; García-Varela and Pérez-Ponce de León, 2008; Presswell et al., 2017). The present different results obtained herein may be related to the different representatives included in the phylogeny or supplementary the ITS sequence data (all the previous studies did not use the ITS sequence data for phylogenetic analyses). However, a more rigorous molecular phylogenetic study including broader representatives of the Polymorphidae using more nuclear and mitochondrial sequence data is required to further clarify the phylogenetic relationships of Southwellina and the other genera of Polymorphidae in the future.
The evolutionary relationships of the polymorphid acanthocephalans revealed by the present molecular analyses enabled us to speculate a possible host-switch pattern (parasitic sequence of changes in definitive host) during the evolution of the polymorphid acanthocephalans: the ancestor of polymorphid acanthocephalans seems to have originally parasitized fish-eating waterfowl in continental habitats (i.e., Southwellina, Ibirhynchus, Hexaglandula, Pseudocorynosoma, Arhythmorhynchus, Polymorphus and Profilicollis), then extended to fish-eating seafowls in brackish water and seawater habitats (i.e., Andracantha), and finally, opportunistically infected marine mammals (i.e., Bolbosoma and Corynosoma). However, this evolutionary issue is still open. We need some more direct evidence to support the present hypothesis.
Declaration of competing interest
The authors declare that they have no conflict of interest.
Ethical approval
The authors declare that they have observed all applicable ethical standards.
Acknowledgements
The authors are grateful to Professor Hideo Hasegawa (Faculty of Medicine, Oita University, Japan) for providing important literature. The authors wish to thank the people of the Aleut community and the National Marine Mammal Laboratory scientists for providing helps during collection of the parasites. This study was supported by the National Natural Science Foundation of China (Grant No. 31872197), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB26000000), the Youth Top Talent Support Program of Hebei Province for Dr. Liang Li.
References
- Alcántar-Escalera F.J., García-Varela M., Vázquez-Domínguez E., Pérez-Ponce de León G. Using DNA barcoding to link cystacanths and adults of the acanthocephalan Polymorphus brevis in central Mexico. Mol. Ecol. Resour. 2013;13:1116–1124. doi: 10.1111/1755-0998.12090. [DOI] [PubMed] [Google Scholar]
- Amin O.M. Review of the genus Polymorphus Luhe, 1911 (acanthocephala: Polymorphidae), with the synonymization of Hexaglandula Petrochenk, 1950 and Subcorynosoma Hoklove, 1967, and a key to the species. Qatar Univ. Sci. J. 1992;12:115–123. [Google Scholar]
- Amin O.M. Classification of the acanthocephala. Folia Parasitol. 2013;60:273–305. doi: 10.14411/fp.2013.031. [DOI] [PubMed] [Google Scholar]
- Amin O.M., Margolis L. Redescription of Bolbosoma capitatum (Acanthocephala: Polymorphidae) from false killer whale off Vancouver Island, with taxonomic reconsideration of the species and synonymy of B. physeteris. J. Helminthol. Soc. Wash. 1998;65:179–188. [Google Scholar]
- Arizono N., Kuramochi T., Kagei N. Molecular and histological identification of the acanthocephalan Bolbosoma cf. capitatum from the human small intestine. Parasitol. Int. 2012;61:715–718. doi: 10.1016/j.parint.2012.05.011. [DOI] [PubMed] [Google Scholar]
- Aznar F.J., Pérez-Ponce de León G., Raga J.A. Status of Corynosoma (Acanthocephala: Polymorphidae) based on anatomical, ecological, and phylogenetic evidence, with the erection of Pseudocorynosoma n. gen. J. Parasitol. 2006;92:548–564. doi: 10.1645/GE-715R.1. [DOI] [PubMed] [Google Scholar]
- Baylis H.A. Parasitic Nematoda and acanthocephala collected in 1925–1927. Discov. Mag. 1929;1:541–560. [Google Scholar]
- Beaver P.C., Otsuji T., Otsuji A., Yoshimura H., Uchikawa R., Sato A. Acanthocephalan, probably Bolbosoma, from the peritoneal cavity of man in Japan. Am. J. Trop. Med. Hyg. 1983;32:1016–1018. doi: 10.4269/ajtmh.1983.32.1016. [DOI] [PubMed] [Google Scholar]
- Costa G., Chubb J.C., Veltkamp C.J. Cystacanths of Bolbosoma vasculosum in the black scabbard fish Aphanopus carbo, oceanic horse mackerel Trachurus picturatus and common dolphin Delphinus delphis from Madeira, Portugal. J. Helminthol. 2000;74:113–120. [PubMed] [Google Scholar]
- Criscuolo A., Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 2010;10:210. doi: 10.1186/1471-2148-10-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delyamure S.L. In: Helminthofauna of Marine Mammals (Ecology and Phylogeny) Delyamure S.L., Skrjabin K.I., editors. AN USSR; Moscow: 1955. p. 518. ([in Russian]) [Google Scholar]
- Dimitrova Z.M., Georgiev B.B. Ardeirhynchus n. g. (Palaeacanthocephala: Polymorphida: Polymorphidae), with a redescription of A. spiralis (Rudolphi, 1809) n. comb. Syst. Parasitol. 1994;29:149–158. [Google Scholar]
- Fonseca M.C.G.D., Knoff M., Felizardo N.N., Torres E.J.L., Di Azevedo M.I.N., Gomes D.C., Clemente S.C.S., Iñiguez A.M. Acanthocephalan parasites of the flounder species Paralichthys isosceles, Paralichthys patagonicus and Xystreurys rasile from Brazil. Rev. Bras. Parasitol. Vet. 2019;28:346–359. doi: 10.1590/S1984-29612019031. [DOI] [PubMed] [Google Scholar]
- Fujita T., Waga E., Kitaoka K., Imagawa T., Komatsu Y., Takanashi K., Anbo F., Anbo T., Katuki S., Ichihara S., Fujimori S., Yamasaki H., Morishima Y., Sugiyama H., Katahira H. Human infection by acanthocephalan parasites belonging to the genus Corynosoma found from small bowel endoscopy. Parasitol. Int. 2016;65:491–493. doi: 10.1016/j.parint.2016.07.002. [DOI] [PubMed] [Google Scholar]
- Fukui T., Morisita T. Contribution to the knowledge of two species of Acanthocephala. Jub. Pro Prof. Sadao Yoshida. 1939;1:137–139. [Google Scholar]
- García-Varela M., Nadler S.A. Phylogenetic relationships of Palaeacanthocephala (Acanthocephala) inferred from SSU and LSU rDNA gene sequences. J. Parasitol. 2005;91:1401–1409. doi: 10.1645/GE-523R.1. [DOI] [PubMed] [Google Scholar]
- García-Varela M., Nadler S.A. Phylogenetic relationships among Syndermata inferred from nuclear and mitochondrial gene sequences. Mol. Phylogenet. Evol. 2006;40:61–72. doi: 10.1016/j.ympev.2006.02.010. [DOI] [PubMed] [Google Scholar]
- García-Varela M., Aznar F.J., Pérez-Ponce de León G., Piñero D., Laclette J.P. Molecular phylogeny of Corynosoma Lühe, 1904 (Acanthocephala), based on 5.8S and internal transcribed spacer sequences. J. Parasitol. 2005;91:345–352. doi: 10.1645/GE-3272. [DOI] [PubMed] [Google Scholar]
- García-Varela M., Pérez-Ponce de León G. Validating the systematic position of Profilicollis Meyer, 1931 and Hexaglandula Petrochenko, 1950 (acanthocephala: Polymorphidae) using cytochrome coxidase (cox 1) J. Parasitol. 2008;94:212–217. doi: 10.1645/GE-1257.1. [DOI] [PubMed] [Google Scholar]
- García-Varela M., Pérez-Ponce de León G., Aznar F.J., Nadler S.A. Systematic position of Pseudocorynosoma and Andracantha (Acanthocephala, Polymorphidae) based on nuclear and mitochondrial gene sequences. J. Parasitol. 2009;95:178–185. doi: 10.1645/GE-1538.1. [DOI] [PubMed] [Google Scholar]
- García-Varela M., Pérez-Ponce de León G., Aznar F.J., Nadler S.A. Erection of Ibirhynchus gen. nov. (Acanthocephala: Polymorphidae), based on molecular and morphological data. J. Parasitol. 2011;97:97–105. doi: 10.1645/GE-2350.1. [DOI] [PubMed] [Google Scholar]
- García-Varela M., Pérez-Ponce de León G., Aznar F.J., Nadler S.A. Phylogenetic relationship among genera of Polymorphidae (Acanthocephala), inferred from nuclear and mitochondrial gene sequences. Mol. Phylogenet. Evol. 2013;68:176–184. doi: 10.1016/j.ympev.2013.03.029. [DOI] [PubMed] [Google Scholar]
- Garey J.R., Near T.J., Nonnemacher M.R., Nadler S.A. Molecular evidence for acanthocephala as a subtaxon of Rotifera. J. Mol. Evol. 1996;43:287–292. doi: 10.1007/BF02338837. [DOI] [PubMed] [Google Scholar]
- Gazi M., Kim J., Park J.K. The complete mitochondrial genome sequence of Southwellina hispida supports monophyly of Palaeacanthocephala (Acanthocephala: Polymorphida) Parasitol. Int. 2015;64:64–68. doi: 10.1016/j.parint.2015.01.009. [DOI] [PubMed] [Google Scholar]
- Gómez A., Serra M., Carvalho G.R., Lunt D.H. Speciation in ancient cryptic species complexes: evidence from the molecular phylogeny of Brachionus plicatilis (Rotifera) Evolution. 2002;56:1431–1444. doi: 10.1111/j.0014-3820.2002.tb01455.x. [DOI] [PubMed] [Google Scholar]
- Guillén-Hernández S., García-Varela M.L., Pérez-Ponce De G. First record of Hexaglandula corynosoma (Travassos, 1915) Petrochenko, 1958 (acanthocephala: Polymorphidae) in intermediate and definitive hosts in Mexico. Zootaxa. 2008;1873:61–68. [Google Scholar]
- Hino Y., Tsuchihashi Y., Kobayashi M., Arizono N., Kagei N. Infection with Bolbosoma sp. (Acanthocephala) in man, report of a case. Clin. Parasitol. 2002;13:102–104. [Google Scholar]
- Ishikura H.A., Takahashi S., Sato N., Kon S., Oku Y., Kamiya M., Ishikura H., Yagi K., Ishii H., Yamamoto H., Kamura T., Kamiya M., Kikuchi K. Perforative peritonitis by the infection with young adult female of Bolbosoma sp.: a case report. Jap. J. Parasitol. 1996;45:518–524. [Google Scholar]
- Isoda K., Kuroda M., Shimizu T., Okumura E. A case of gastric acanthocephalan infection, Bolbosoma sp., found by gastroendoscopy. Clin. Parasitol. 2006;17:83–88. [Google Scholar]
- Kaito S., Sasaki M., Goto K., Matsusue R., Koyama H., Nakao M., Hasegawa H. A case of small bowel obstruction due to infection with Bolbosoma sp. (Acanthocephala: Polymorphidae) Parasitol. Int. 2019;68:14–16. doi: 10.1016/j.parint.2018.09.007. [DOI] [PubMed] [Google Scholar]
- Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., Von Haeseler A., Jermiin L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Li L. First report on cystacanths of Sphaerirostris lanceoides (Petrochenko, 1949) (acanthocephala: Centrorhynchidae) from the Asiatic toad Bufo gargarizans Cantor (Amphibia: Anura) in China. Syst. Parasitol. 2018;95:447–454. doi: 10.1007/s11230-018-9794-0. [DOI] [PubMed] [Google Scholar]
- Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Král’ová-Hromadová I., Tietz D.F., Shinn A.P., Spakulová M. ITS rDNA sequences of Pomphorhynchus laevis (Zoega in Müller, 1776) and P. lucyi Williams and Rogers, 1984 (acanthocephala: Palaeacanthocephala) Syst. Parasitol. 2003;56:141–145. doi: 10.1023/a:1026127219358. [DOI] [PubMed] [Google Scholar]
- Kuzmina T.A., Kuzmin Y., Dzeverin I., Lisitsyna O.I., Spraker T.R., Korol E.M., Kuchta R. Review of metazoan parasites of the northern Fur seal (Callorhinus ursinus) and the analysis of the gastrointestinal helminth community of the population on St. Paul Island, Alaska. Parasitol. Res. 2021;120:117–132. doi: 10.1007/s00436-020-06935-6. [DOI] [PubMed] [Google Scholar]
- Kuzmina T.A., Lisitsyna O.I., Lyons E.T., Spraker T.R., Tolliver S.C. Acanthocephalans in northern Fur seals (Callorhinus ursinus) and a harbor seal (Phoca vitulina) on St. Paul Island, Alaska: species, prevalence, and biodiversity in four Fur seal subpopulations. Parasitol. Res. 2012;111:1049–1058. doi: 10.1007/s00436-012-2930-x. [DOI] [PubMed] [Google Scholar]
- Letunic I., Bork P. Interactive Tree of Life (ITOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021;49:293–296. doi: 10.1093/nar/gkab301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Wayland M.T., Chen H.X., Yang Y. Remarkable morphological variation in the proboscis of Neorhadinorhynchus nudus (Harada, 1938) (acanthocephala: Echinorhynchida) Parasitology. 2019;146:348–355. doi: 10.1017/S003118201800166X. [DOI] [PubMed] [Google Scholar]
- Lisitsyna O.I., Kudlai O., Spraker T.R., Tkach V.V., Smales L.R., Kuzmina T.A. Morphological and molecular evidence for synonymy of Corynosoma obtuscens Lincicome, 1943 with Corynosoma australe Johnston, 1937 (acanthocephala: Polymorphidae) Syst. Parasitol. 2019;96:95–110. doi: 10.1007/s11230-018-9830-0. [DOI] [PubMed] [Google Scholar]
- Malyarchuk B., Derenko M., Mikhailova E., Denisova G. Phylogenetic relationships among Neoechinorhynchus species (Acanthocephala: Neoechinorhynchidae) from North-East Asia based on molecular data. Parasitol. Int. 2014;63:100–107. doi: 10.1016/j.parint.2013.09.012. [DOI] [PubMed] [Google Scholar]
- Minh B.Q., Schmidt H.A., Chernomor O., Schrempf D., Woodhams M.D., Von Haeseler A., Lanfear R. IQ-TREE 2: new models and efficient methods for phylogenetic inference in the Genomic Era. Mol. Biol. Evol. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori S., Maeba T., Sekimata T., Kobayashi S., Suguri S., Harada M., Kagei N. A human infection with Bolbosoma sp., accidently found in the operation for gastric cancer. Clin. Parasitol. 1998;9:35–37. [Google Scholar]
- Muhammad N., Suleman, Khan M.S., Li L., Zhao Q., Ullah H., Zhu X.Q., Ma J. Characterization of the complete mitogenome of Centrorhynchus clitorideus (Meyer, 1931) (Palaeacanthocephala: Centrorhynchidae), the largest mitochondrial genome in Acanthocephala, and its phylogenetic implications. Mol. Biochem. Parasitol. 2020;237 doi: 10.1016/j.molbiopara.2020.111274. [DOI] [PubMed] [Google Scholar]
- Near T.J., Garey J.R., Nadler S.A. Phylogenetic relationships of the Acanthocephala inferred from 18S ribosomal DNA sequences. Mol. Phylogenet. Evol. 1998;10:287–298. doi: 10.1006/mpev.1998.0569. [DOI] [PubMed] [Google Scholar]
- Nguyen L.T., Schmidt H.A., Von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presswell B., García-Varela M., Smales L.R. Morphological and molecular characterization of two new species of Andracantha (Acanthocephala: Polymorphidae) from New Zealand shags (Phalacrocoracidae) and penguins (Spheniscidae) with a key to the species. J. Helminthol. 2017;92:740–751. doi: 10.1017/S0022149X17001067. [DOI] [PubMed] [Google Scholar]
- Ronquist F., Teslenko M., Van der Mark P., Ayres D.L., Darling A., Höhna S., Larget B., Liu L., Suchard M.A., Huelsenbeck J.P. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro M., Palomba M., Gili C., Marcer F., Marchiori E., Mattiucci S. Molecular and morphological characterization of Bolbosoma balaenae (Acanthocephala: Polymorphidae), a neglected intestinal parasite of the fin whale Balaenoptera physalus. Parasitology. 2021;148:1293–1302. doi: 10.1017/S0031182021000925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki M., Katahira H., Kobayashi M., Kuramochi T., Matsubara H., Nakao M. Infection status of commercial fish with cystacanth larvae of the genus Corynosoma (Acanthocephala: Polymorphidae) in Hokkaido, Japan. Int. J. Food Microbiol. 2019;305 doi: 10.1016/j.ijfoodmicro.2019.108256. [DOI] [PubMed] [Google Scholar]
- Schmidt G.D. Andracantha, a new genus of Acanthocephala (Polymorphidae) from fish-eating birds, with descriptions of three species. J. Parasitol. 1975;61:615–620. [PubMed] [Google Scholar]
- Skrjabin A.S. Morphological differences between Bolbosoma turbinella (Diesing, 1851) (fam. Polymorphidae) acanthocephala from the north and south hemispheres. Parazitologia. 1972;6:57–64. [PubMed] [Google Scholar]
- Steinauer M.L., Nickol B.B., Ortí G. Cryptic speciation and patterns of phenotypic variation of a highly variable acanthocephalan parasite. Mol. Ecol. 2007;16:4097–4109. doi: 10.1111/j.1365-294X.2007.03462.x. [DOI] [PubMed] [Google Scholar]
- Tada I., Otsuji Y., Kamiya H., Mimori T., Sakaguchi Y., Makizumi S. The first case of a human infected with an acanthocephalan parasite, Bolbosoma sp. J. Parasitol. 1983;69:205–208. [PubMed] [Google Scholar]
- Verweyen L., Klimpel S., Palm H.W. Molecular phylogeny of the Acanthocephala (class Palaeacanthocephala) with a paraphyletic assemblage of the orders Polymorphida and Echinorhynchida. PLoS One. 2011;6 doi: 10.1371/journal.pone.0028285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waindok P., Lehnert K., Siebert U., Pawliczka I., Strube C. Prevalence and molecular characterisation of acanthocephala in pinnipedia of the north and Baltic Seas. Int. J. Parasitol-Par. 2018;7:34–43. doi: 10.1016/j.ijppaw.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P.Q. Notes on the acanthocephala from Fujlan Ⅱ. Acta Zootaxonomica Sin. 1980;5:116–123. [Google Scholar]
- Wayland M.T., Vainio J.K., Gibson D.I., Herniou E.A., Littlewood D.T.J., Väinölä R. The systematics of Echinorhynchus Zoega in Müller, 1776 (Acanthocephala, Echinorhynchidae) elucidated by nuclear and mitochondrial sequence data from eight European taxa. ZooKeys. 2015;484:25–52. doi: 10.3897/zookeys.484.9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguti S. Studies on the helminth fauna of Japan. Part 8. Acanthocephala I. Jap. J. Zool. 1939;8:317–351. [Google Scholar]
- Zhang D., Gao F., Jakovlić I., Zou H., Zhang J., Li W.X., Wang G.T. PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020;20:348–355. doi: 10.1111/1755-0998.13096. [DOI] [PubMed] [Google Scholar]
- Zhao Q., Muhammad N., Chen H.X., Ma J., Suleman Li L. Morphological and genetic characterization of Centrorhynchus clitorideus (Meyer, 1931) (acanthocephala: Centrorhynchidae) from the little owl Athene noctua (Scopoli) (Strigiformes: Strigidae) in Pakistan. Syst. Parasitol. 2020;97:517–528. doi: 10.1007/s11230-020-09930-8. [DOI] [PubMed] [Google Scholar]
- Zittel M., Grabner D., Wlecklik A., Sures B., Leese F., Taraschewski H., Weigand A.M. Cryptic species and their utilization of indigenous and non-indigenous intermediate hosts in the acanthocephalan Polymorphus minutus sensu lato (Polymorphidae) Parasitology. 2018;145:1421–1429. doi: 10.1017/S0031182018000173. [DOI] [PubMed] [Google Scholar]