FIGURE 6.
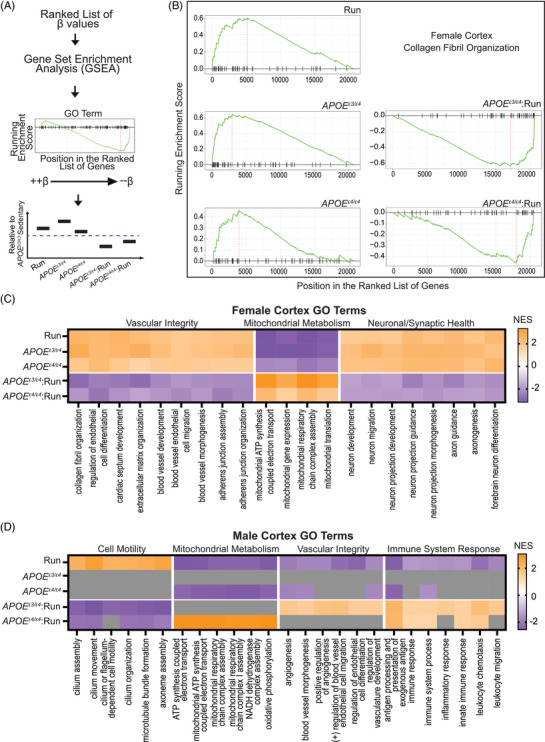
Gene set enrichment analysis (GSEA) predicts apolipoprotein E (APOE) genotype and running interact to mitigate main effects. A, Schematic of computational approach; β‐values from linear models were passed through GSEA for gene ranking, GSEA plots were used to visualize results and main effects of running, APOE genotype, and APOE genotype:running were interpreted per Gene Ontology (GO) term. B, GSEA plots for “Collagen Fibril Organization” in the female cortex. Main effects of running and APOE ε3/ε4, and APOE ε4/ε4 all show positive enrichment scores, while the interactions, APOE ε3/ε4:run and APOE ε4/ε4:run reveal negative enrichment scores. C, In the female cortex data GO terms that fit the pattern shown in (B), colored by normalized enrichment score (NES), are represented specifically vascular integrity, mitochondrial metabolism, and synaptic/neuronal health. D, The pattern observed in male cortex was different than that seen in female cortex (B,C) with APOE ε3/ε4 appearing more similar to APOE ε3/ε3 (indicated by gray boxes) for enrichment terms grouped as cell motility, mitochondrial metabolism, vascular integrity, and immune system response
