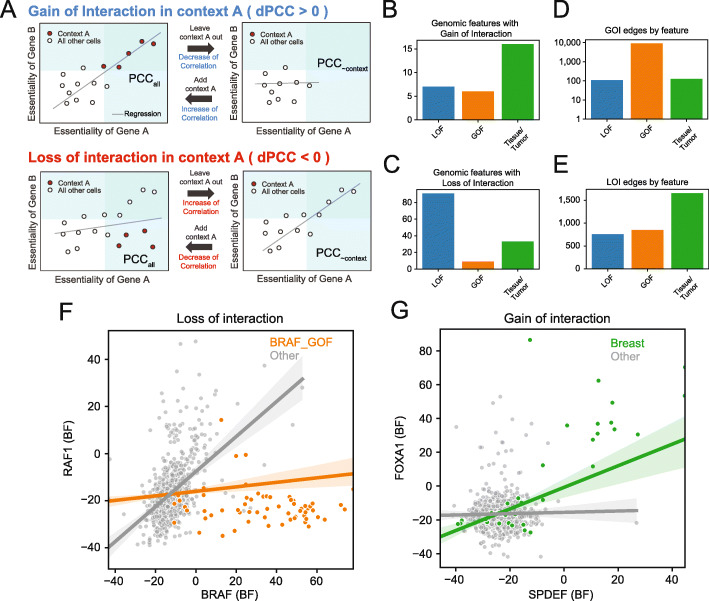
Fig. 2.
A framework to identify dynamics in coessentiality network. A Classes of context-dependent interaction. In all cases, a differential Pearson correlation coefficient (dPCC) for each gene pair is calculated as the difference between the global network (PCCall) and the PCC from all cells except those harboring the feature of interest (PCC~context). B, C The number of features associated with gain or loss of interaction. D, E the number of gained or lost interactions, by feature type. Nearly all GOI edges are associated with TP53_GOF mutations. F, G Examples of context-dependent loss and gain of interaction. Axes are Bayes Factors of the genes