Extended Data Fig. 1. Generation of RNA transcript standards for validation of SARS-CoV-2 RT-qPCR assays.
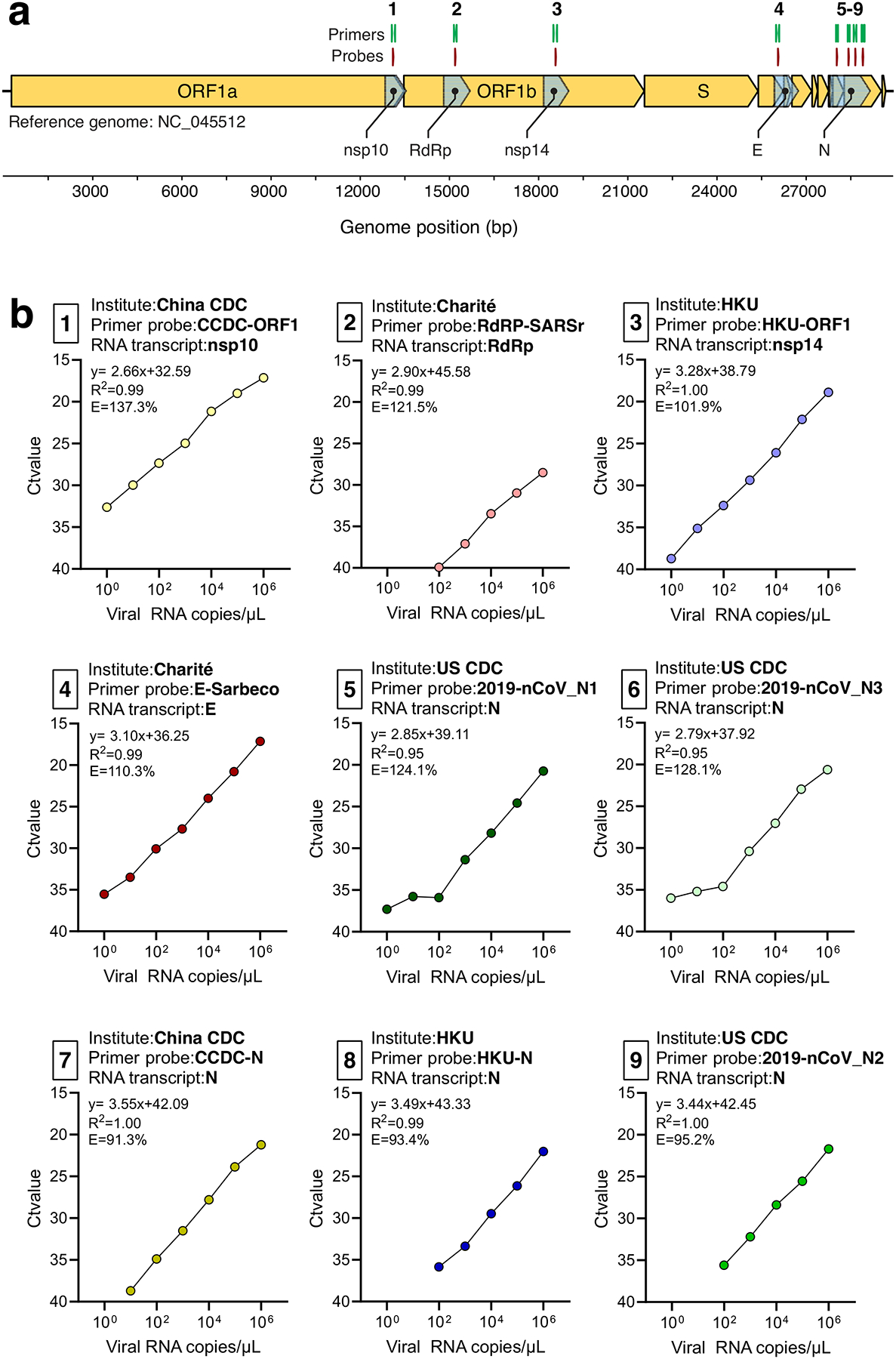
a, SARS-CoV-2 genome locations of generated RNA transcript standards for the non-structural protein 10 (nsp10), RNA-dependent RNA polymerase (RdRp), non-structural protein 14 (nsp14), envelope (E), and nucleocapsid (N) genes and the nine primer-probe sets used RT-qPCR assays. b, The slope, intercept, R2, and efficiency of RT-qPCR using tenfold dilutions (100-106 viral RNA copies/μL) of RNA transcript standards with the corresponding primer-probe sets. Shown are mean Ct values based on 2 technical replicates. The primer-probe sets are numbered as shown in panel A. The RNA transcript primers and sequences can be found in Supplementary Table 2 and Supplementary Table 3, respectively. Data used to make this figure can be found in Source Data Extended Data Fig. 1.
