Summary
The cultivation of rice varieties with high seed vigour is vital for the direct seeding of rice, and the molecular basis of regulation of seed vigour remains elusive. Here, we cloned a new gene OsHIPL1, which encodes hedgehog‐interacting protein‐like 1 protein as a causal gene of the major QTL qSV3 for rice seed vigour. OsHIPL1 was mainly localized in the plasma membrane and nucleus. RNA sequencing (RNA‐seq) revealed that the ABA‐related genes were involved in the OsHIPL1 regulation of seed vigour in rice. The higher levels of endogenous ABA were measured in germinating seeds of OsHIPL1 mutants and NIL‐qsv3 line compared to IR26 plants, with two up‐regulated ABA biosynthesis genes (OsZEP and OsNCED4) and one down‐regulated ABA catabolism gene OsABA8ox3. The expression of abscisic acid‐insensitive 3 (OsABI3), OsABI4 and OsABI5 was significantly up‐regulated in germinating seeds of OsHIPL1 mutants and NIL‐qsv3 line compared to IR26 plants. These results indicate that the regulation of seed vigour of OsHIPL1 may be through modulating endogenous ABA levels and altering OsABIs expression during seed germination in rice. Meanwhile, we found that OsHIPL1 interacted with the aquaporin OsPIP1;1, then affected water uptake to promote rice seed germination. Based on analysis of single‐nucleotide polymorphism data of rice accessions, we identified a Hap1 haplotype of OsHIPL1 that was positively correlated with seed germination. Our findings showed novel insights into the molecular mechanism of OsHIPL1 on seed vigour.
Keywords: rice, seed vigour, OsHIPL1, ABA signalling, OsPIP1;1
Introduction
Rice (Oryza sativa L.) is the staple food resource for half of the world's population. Rice direct seeding has been popularized because of its lower labour costs compared to the conventional transplanting of seedlings (Kumar and Ladha, 2011; Liu et al., 2015). Seed germination and seedling establishment as key events in plant life activities are the important factors influencing the yield of direct seeding in rice (Gommers and Monte, 2018; Rajjou et al., 2012). In the production fields of rice direct seeding, seed germination and seedling establishment could be delayed due to adverse environmental factors from paddy water surface or puddled soils, greatly increasing seedling mortality with a high risk of yield loss (Mahender et al., 2015; Rao et al., 2007). Seeds with high vigour have the power potential for rapid, uniform germination and the development of strong seedlings in rice fields, with the inhibition of weed growth (Foolad et al., 2007; Wang et al., 2010). Thus, the improvement of seed vigour is essential for the direct seeding of rice (Mahender et al., 2015).
Seed vigour is a complex quantitative trait that is determined by the interactions between genetic and environmental factors. Many quantitative trait loci (QTL) for seed vigour, such as germination potential, germination index, germination percentage and seedling percentage, have been identified using the biparental mapping approach in rice (Fujino et al., 2004, 2008; He et al., 2019; Jiang et al., 2011, 2017; Liu et al., 2014; Xie et al., 2014; Zeng et al., 2021). Given these QTLs, some of the genes have been cloned and elucidated for seed vigour in rice. The major qLTG3‐1 has been map‐based cloned and its expression is tightly associated with vacuolation of the tissues covering the embryo during seed germination (Fujino et al., 2008). qSE3 encoding a K+ transporter OsHAK21 was isolated to promote rice seed germination and seedling establishment under salinity stress (He et al., 2019). Therefore, it is helpful to improve seed vigour of rice in breeding programme through further exploring and cloning the seed vigour‐related genes, and elucidating their molecular mechanisms.
Viable seeds can rapidly germinate at favourable conditions, and its radicle protrudes the seed coats with growth of the seedlings (Bewley, 1997). Seed germination and seedling establishment are two important features that reflect the capacities of seed vigour to some extent. The process of seed germination is involved in a series of coordinated physiological and biochemical initiations, including the increase in oxygen and water uptake, the activation of the glycolysis, pentose phosphate pathway and tricarboxylic acid (TCA) cycle (He and Yang, 2013; Rajjou et al., 2012). Previous studies show that well‐germinated seeds are closely associated with the balance between internal reactive oxygen species (ROS) contents and the activities of ROS scavenging systems (Bailly et al., 2008; El‐Maarouf‐Bouteau and Bailly, 2008; He et al., 2019). High abundance of late embryogenesis abundant (LEA) protein and aquaporins has significant contributions to the hydraulic activity of cells and re‐establishment of metabolism during seed germination (Footitt et al., 2019; Vander Willigen et al., 2006). The plant hormones, such as abscisic acid (ABA), gibberellin (GA) and auxin (IAA), also take crucial roles in the regulation in cell homeostasis for seed germination and seedling establishment (Abe et al., 2012; Debeaujon and Koornneef, 2000; Li et al., 2016; Shu et al., 2016).
It is well known that ABA as the major endogenous factor has been widely investigated involving the inhibition of seed germination (Penfield, 2017). The ABA levels in plants are maintained in strict homeostasis, including biosynthesis, degradation and conjugation, during various physiological processes (Palaniyandi et al., 2015; Seiler et al., 2011). The increase in ABA levels in seeds is controlled by ABA biosynthesis genes zeaxanthin epoxidase (ZEP) and 9‐cis‐epoxycaretonoid dioxygenases (NCEDs) (Eiji et al., 2010). The decrease in ABA levels is regulated by ABA catabolism CYP707A family genes (Kushiro et al., 2004). ABA content increases in seeds during the seed maturation and dormancy induction processes to activate the ABA signalling pathway (Shu et al., 2016). ABA signalling is perceived by a set of receptors, which are members of the PYR1/PYL/RCAR family (Ma et al., 2009; Park et al., 2009). ABA binding to PYR1/PYL/RCAR complex leads to deactivation of protein phosphatase 2C, which releases and activates SnRK2 kinases (Chen et al., 2020; Cutler et al., 2010). Activated SnRK2s can phosphorylate ABA‐responsive element (ABRE)‐binding factors (ABFs) that promote the expression of ABA‐dependent genes (Chen et al., 2020; Sano and Marion‐Poll, 2021; Song et al., 2021). Among the identified ABF TFs (transcription factors), the B3 domain TF ABI3, the APETALA2 domain TF ABI4 and the basic leucine zipper (bZIP) TF ABI5 play central roles in regulating seed germination and early seedling growth (Ding et al., 2014; Lopez‐Molina et al., 2001; Luo et al., 2021). Loss‐of‐function abi3 conferred reduced sensitivity to ABA during seed germination and early seedling growth (Ding et al., 2014). ABI4 can directly combine with RbohD and vitamin C defective 2 (VTC2) and promotes ROS accumulation to decrease seed germination under salinity stress in Arabidopsis (Luo et al., 2021). The germination of abi5 mutants is insensitive to ABA, while ABI5‐overexpressing plants are allergic to ABA (Finkelstein and Lynch, 2000; Lopez‐Molina et al., 2001). In rice, OsIAGLU, an indole‐3‐acetate beta‐glucosyltransferase gene, was involved in the regulation of seed vigour through modulated OsABIs expressions in germinating seeds (He et al., 2020).
Hedgehog‐interacting protein (HIP) was identified as a hedgehog (Hh)‐binding protein that is a target of Hh signalling (Chuang and McMahon, 1999). The Hh signalling pathway is essential for the development of diverse tissues during embryogenesis (Hammerschmidt et al., 1997). Hh signalling is activated by binding of hedgehog protein to the multipass membrane protein Patched (Ptc) (Tabin and McMahon, 1997). Loss‐of‐function mutants in HIP1 result in an up‐regulation of Hh signalling in the mouse embryo, disrupting cell interactions essential for the normal morphogenesis of the lung and skeleton (Chuang et al., 2003). Recently, the hedgehog‐interacting protein‐like 1 (HHIPL1) in human was reported as a secreted proatherogenic protein that enhances Hh signalling and regulates smooth muscle cell proliferation and migration (Aravani et al., 2019). In Arabidopsis, it was reported that there were two hedgehog‐interacting protein‐like protein (HIPLs) belonging to glycosyl‐phosphatidyl‐inositol (GPI)‐anchored proteins (GAPs) (Borner et al., 2003). GAPs are involved in the generation of specialized cell surfaces and extracellular signalling molecules, likely targeting a specific subset of proteins to the cell surface for extracellular matrix remodelling and signalling (Borner et al., 2002; Sherrier et al., 1999). These imply that the HIPL might have potential roles in plants.
In our previous studies, a chromosome segment substitution lines (CSSL) population was structured based on the recipient parent IR26 and donor Jiucaiqing (JCQ) (Cheng et al., 2016; He et al., 2019). Here, we conducted QTL mapping of seed vigour using the CSSL population, and found a major locus qSV3 on chromosome 3 improving seed germination and seedling establishment. The genetic and transgenic information showed that OsHIPL1, encoding a hedgehog‐interacting protein‐like 1 protein, was the causal gene of qSV3. Our results suggest that OsHIPL1 acts in the regulation of seed vigour through the ABA pathway in seed germination. Moreover, we observed that OsHIPL1 interacted with OsPIP1;1 in rice. The application of qSV3 may be useful in rice breeding programmes for improvement of rice seed vigour for direct seeding cultivation.
Results
Map‐based cloning of qSV3
According to the previous report (He et al., 2019), indica rice cultivar IR26 showed significantly high seed vigour under H2O condition compared with japonica Jiucaiqing (Figures 1a and S1). To identify the elite genes controlling high seed vigour in IR26, a population consisting of 62 CSSLs was used for QTL identification and mapping. The seed germination percentage (GP), germination index (GI) and seedling percentage (SP) of 62 CSSLs, as well as their two parents, were assessed under H2O condition respectively (Figure 1a, Tables S1 and S2). By single marker analysis (SMA), we identified seven QTLs, qGP3.1, qGI3.1, qGP3.2, qGI3.2, qGP10.1, qSP3.1 and qSP10.1, for GP, SP and GI, respectively, and their positive alleles were derived from IR26 with an increase in seed vigour, except qGP3.1 and qGI3.1 (Figure 1b, Table S3). The qGP3.2, qGI3.2 and qSP3.1 were mapped to be tightly associated with marker RM3513 on chromosome 3, integrated as a qSV3 locus (Table S3). The qGP10.1 and qSP10.1 were mapped to be tightly associated with the marker RM5348 on chromosome 10, integrated as a qSV10 locus (Table S3). Since the qSV3 showed higher contribution to seed vigour with 37.75% of average phenotypic variation (APV) of GP, 21.74% of APV of SP and 88.52% of APV of GI, it was further analysed in this study.
Figure 1.
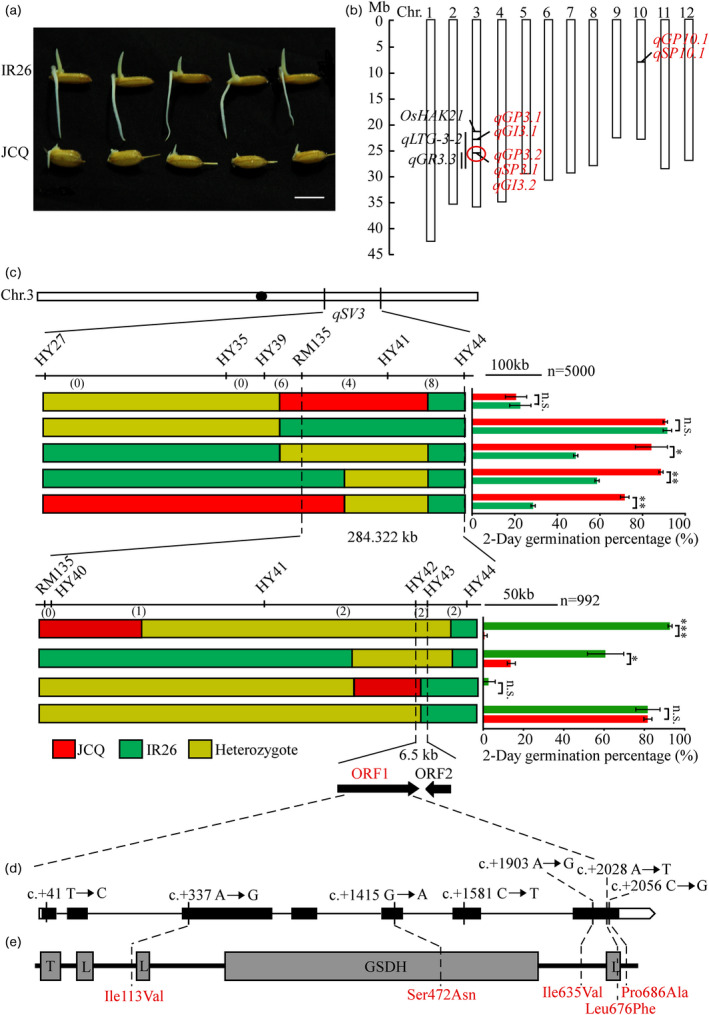
Map‐based cloning of qSV3. (a) Parental phenotype after 3 days after germination. Bar = 1 cm. (b) Physical mapping of QTLs related to seed vigour. Red QTLs identified based on CSSL population in this study and circle locus includes the major qGP3.2, qGI3.2 and qSP3.1, which is named qSV3. The black QTLs have been reported previously and black lines show their physical regions. (c) The qSV3 locus was fined within a 6.5 kb region between markers HY42 and HY43 on chromosome 3. Black circle represents the centromere. Numbers below the horizontal line are the number of recombinants. Red bars represent the JCQ genotype region; green and yellow bars represent IR26 genotype and heterozygous genotype respectively. Right: Germination percentage 2 days after germination. Each column presents the means ± standard deviations of three biological replicates (*P < 0.05, **P < 0.01 and ***P < 0.001) compared with the control by Student’s t‐test. n.s. represents no significance. Candidate genes ORF1 and ORF2 are LOC_Os03g48540 and LOC_Os03g48550 respectively. (d) Gene structure of ORF1. Empty boxes refer to 5′ and 3′ UTRs, black boxes to exons and the lines between boxes to introns. The SNPs from JCQ to IR26 in the OsHIPL1 are shown by solid lines; SNP1, c.+41T→C; SNP2, c.+337A→G; SNP3, c.+1415G→A; SNP4, c.+1581C→T; SNP5, c.+1903A→G; SNP6, c.+2028A→T; and SNP7, c.+2056C→G. (e) Protein structure of ORF1. T, L and GSDH represents the transmembrane domain, the low complexity region and glucose/sorbosone dehydrogenase domain predicted respectively. Dashed lines show the positions of two amino acid transitions.
The residual heterozygous line CSSL10, containing the allele (qsv3) from parent Jiucaiqing (Figure S2A, Table S1) with the low GP, SP and GI, was selected for confirming qSV3 locus. A segregation population consisting of 120 individuals was developed by self‐fertilization of CSSL10, and their GP at 2 days after imbibition showed bimodal characteristic distribution (Figure S2B), suggesting the presence of a major gene. Inclusive composite interval mapping (ICIM) showed that there was a significant QTL of GP between markers HY27 and HY44, with 55.48% of phenotypic variation (Figure S2C). A total of 5000 individuals from the BC6F4 population were used to narrow down the qSV3 locus into a genomic region between the markers RM135 and HY44 (Figure 1c). A total of 992 individuals of the BC6F5 population were used to further delimit the qSV3 locus in an approximately 6.5 kb region between the markers HY42 and HY43 (Figure 1c).
In this region, two genes, LOC_Os03g48540 (ORF1) and LOC_Os03g48550 (ORF2), were predicted in the Rice Annotation Project Database (http://rice.uga.edu/index.shtml). ORF1 encodes a hedgehog‐interacting protein‐like 1 protein, designated as OsHIPL1, with seven exons and six introns, and ORF2 encodes one retrotransposon protein. To define the candidate gene of qSV3, the cDNA of these two ORFs was cloned from IR26 and Jiucaiqing, respectively, and sequenced for comparison. There were no nucleotide differences in ORF2 between IR26 and Jiucaiqing (Figure S3), but seven single‐nucleotide polymorphisms (SNPs) (SNP1–SNP7) were found in ORF1 (OsHIPL1) (Figure 1d). Among these SNPs, five SNPs caused amino acid residue changes between IR26 and Jiucaiqing. SNP2 in the third exon of OsHIPL1 caused amino acid residue change from isoleucine in IR26 to valine in Jiucaiqing (Ile1137Val); SNP3 in the fifth exon of OsHIPL1 caused serine to asparagine (Ser472Asn); and SNP5, SNP6 and SNP7 in the seventh exon of OsHIPL1 caused isoleucine to valine (Ile635Val), leucine to phenylalanine (Lue676Phe) and proline to alanine (Pro686Ala) respectively (Figure 1d–e).
SMART (http://smart.embl‐heidelberg.de/) results showed that OsHIPL1 contains 699 amino acids with one transmembrane domain, three low complexity regions and one glucose/sorbosone dehydrogenase (GSDH) domain. SNP3 and SNP6 were predicted to locate in GSDH domain and the third low complexity region respectively (Figure 1e). Thus, OsHIPL1 was considered as a causal gene for qSV3.
Confirmation of OsHIPL1 as qSV3
Three homozygous knockout mutants (Oshipl1a, Oshipl1b and Oshipl1c, T2) were developed in the background of IR26 through the CRISPR/Cas9 system (Figure 2a). The Oshipl1a contained a ‘T’ and an ‘A’ insertion in the first and second exons of OsHIPL1, respectively; the Oshipl1b, an ‘A’ insertion in the first and second exons of OsHIPL1, respectively; and the Oshipl1c, a ‘27bp’ deletion and a ‘C’ insertion in the first and second exons of OsHIPL1 respectively (Figure 2a). Based on these nucleotide sequences, the amino acid sequence of OsHIPL1 was predicted to contain only 64, 9 and 85 amino acids in Oshipl1a, Oshipl1b and Oshipl1c mutants, respectively, which were caused by premature termination (Figure 2b). This result indicates that the mutants lacked OsHIPL1 gene. Germination assays showed that the GP, SP and GI of Oshipl1a, Oshipl1b and Oshipl1c mutants were significantly reduced compared with those of IR26 (Figure 2c–f), suggesting that the disruptions of OsHIPL1 gene significantly decreased seed vigour.
Figure 2.

Functional analysis of OsHIPL1 during seed germination. (a) Target sites are marked in red. Sequences in boxes are the target sequences in the wild type (WT). Black dashes represent the deleted bases and inserted nucleotides are indicated with red uppercase letters. (b) Analysis of the OsHIPL1 protein sequence in the wild‐type (WT) and OsHIPL1‐knockout lines. (c) Photographs of germinated seeds of Oshipl1 mutants and IR26 (control) after 3 days after germination. (d–f) Comparison of germination percentage (d), seedling percentage (e) and germination index (f) between Oshipl1 mutants and IR26. (g) Photographs of germinated seeds of COM lines and NIL‐qsv3 (control) after 3 days after germination. (h–j) Comparison of germination percentage (h), seedling percentage (i) and germination index (j) between COM lines and NIL‐qsv3. (k) Photographs of germinated seeds of OE lines and ZH11 (control) after 3 days after germination. (l–n) Comparison of germination percentage (l), seedling percentage (m) and germination index (n) between OE lines and ZH11. Bar = 1 cm. Each column presents the means ± standard deviations of three biological replicates. *P < 0.05, **P < 0.01 and ***P < 0.001 compared with the control by Student’s t‐test.
A near isogenic line (NIL), containing a small chromosomal segment around qsv3 from Jiucaiqing (NIL‐qsv3), was developed in IR26 genetic background. The NIL‐qsv3 showed a lower seed vigour than IR26 (Figure S4). To further confirm whether OsHIPL1 is the causal gene of qSV3, we performed a genetic complementation test in the NIL‐qsv3 background. The OsHIPL1 expression levels in three complementation lines (COM‐1, COM‐2 and COM‐3, T2) were 7‐fold, 9‐fold and 10‐fold higher than those in NIL‐qsv3 respectively (Figure S5A). Germination assays showed that the GP, SP and GI of complementation lines were significantly increased, compared with NIL‐qsv3 (Figure 2g–j), indicating that the allele (OsHIPL1) of IR26 compensates the defective function of qsv3 allele on seed vigour.
We further generated OsHIPL1 overexpression lines (OE‐1, OE‐2 and OE‐3, T2) containing the 35S promoter fused to the OsHIPL1‐coding region from IR26 transformed into japonica Zhonghua 11 (ZH11). The OsHIPL1 expression levels in OE‐1, OE‐2 and OE‐3 lines were 9‐fold, 13‐fold and 17‐fold higher than that in ZH11, respectively (Figure S5B). Seed germination assays showed that the GP, SP and GI of the overexpression lines were significantly increased compared with those of ZH11 (Figure 2k–n). It indicates that OsHIPL1 is a positive regulator for seed vigour in rice.
Expression patterns of OsHIPL1 and subcellular localization
By the quantitative RT‐PCR (RT‐qPCR) approach, the expression patterns of the OsHIPL1 gene were analysed in various tissues, and germinating and developing seeds in IR26. There was a relatively higher expression of OsHIPL1 identified in the root, compared with that in the stem, leaf, sheath, internode and spike (Figure 3a). During seed germination,, the transcript levels of OsHIPL1 were gradually increased at the early stage (0–30 h after imbibition), then decreased at the late stage (36–72 h after imbibition) (Figure 3b). During seed development, the expression levels of OsHIPL1 gradually increased from 0 to 35 days after flowering (Figure 3c). The specific expression of OsHIPL1 suggested that it might play a role in modulating seed vigour in rice.
Figure 3.

Expression patterns of OsHIPL1 and subcellular localization in rice. Expression pattern of OsHIPL1 in various tissues (a), germinating stages (b) and seed developmental stages (c) in IR26 using the RT‐qPCR approach. The expression of OsHIPL1 was normalized to that of OsActin gene control. The relative expression levels were represented by fold change relative to the expression level of OsHIPL1 at 0 days after flowering or 0 h imbibition stage under H2O treatment. Each column represents the means ± standard deviation. (d) Subcellular localization of OsHIPL1 in N. benthamiana leaves. (e) Subcellular localization of OsHIPL1 tagged at the C terminus with GFP in rice protoplasts.
To determine the subcellular localization of OsHIPL1, a recombinant OsHIPL1 protein‐tagged green fluorescent protein (GFP) at the C terminus under the control of the 35S promoter was constructed and expressed transiently in rice protoplasts and N. benthamiana leaves. The GFP‐tagged OsHIPL1 was found to be mainly localized in the plasma membrane and nucleus in rice protoplasts and N. benthamiana leaf epidermal cells (Figure 3d–e).
OsHIPL1 is involved in ABA‐mediated processes possibly during seed germination
To explore the regulatory mechanism of OsHIPL1 on seed vigour, the RNA‐seq analysis was performed between IR26 and NIL‐qsv3 at the 12th hour (h) after imbibition. A total of 3186 differently expressed genes (DEGs) were identified, including 1432 up‐regulation and 1754 down‐regulation, in NIL‐qsv3 compared to IR26 (P‐value < 0.05, fold change ≥2) (Table S4). Twelve DEGs, including five ABA‐related genes, six IAA‐related genes and one ascorbate peroxidase gene, were randomly selected for their expression by the RT‐qPCR approach in germinating seed of IR26 and NIL‐qsv3. It was observed that there was a consistent expression pattern for those DEGs by the RNA‐Seq and RT‐qPCR approach in germinating seeds between IR26 and NIL‐qsv3 seeds (Figure S6).
All DEGs were analysed based on the gene ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG). GO results showed that the pathways involved in abiotic stimulus in biological process (BP), DNA binding in molecular function (MF) and cell periphery in cellular component (CC) were significantly enriched (Figure S7). KEGG results showed that many DEGs between IR26 and NIL‐qsv3 were involved in the plant hormone signal transduction including ABA, IAA, GA, brassinosteroid (BR), ethylene (ET), cytokinin (CK) and jasmonic acid (JA) (Figure S8A, Table S5).
ABA plays a crucial role in the control of seed dormancy and germination (Finkelstein et al., 2008; Rajjou et al., 2012). Our RNA‐Seq showed that 18 DEGs were related to ABA metabolism, including ABA signal transduction genes (12), ABA response genes (4), ABA biosynthesis gene (1) and ABA inducible gene (1) (Figure S8B). To examine whether the function of OsHIPL1 is dependent on the ABA, we investigated the expression levels of OsHIPL1 under ABA treatments during seed germination. The RT‐qPCR analysis indicated that OsHIPL1 expression was significantly induced by ABA during seed imbibition (Figure S9). These results suggest that the regulation of OsHIPL1 on seed vigour may be involved in ABA‐mediated processes during seed germination in rice.
OsHIPL1 regulates ABA metabolism and signalling during seed germination
To investigate whether OsHIPL1 is involved in ABA responses, we investigated the IR26 and two mutants Oshipl1a and Oshipl1b in response to exogenous ABA (0, 1 and 3 μm) during seed germination. In the presence of ABA, the seed vigour of Oshipl1 mutants and IR26 was significantly reduced (Figure 4a–d), and the seed vigour of Oshipl1 mutants declined more than that of IR26 plants. Furthermore, we investigated the COM‐1, COM‐2 and NIL‐qsv3 lines in response to exogenous ABA (0, 1 and 3 μm) during seed germination. In the presence of ABA, the seed vigour of complementation and NIL‐qsv3 lines were significantly reduced. Moreover, the seed vigour of the complementation lines declined less than that of NIL‐qsv3 line (Figure 4e–h). This suggests that the function of OsHIPL1 can relieve the inhibited effect of exogenous ABA during seed germination.
Figure 4.
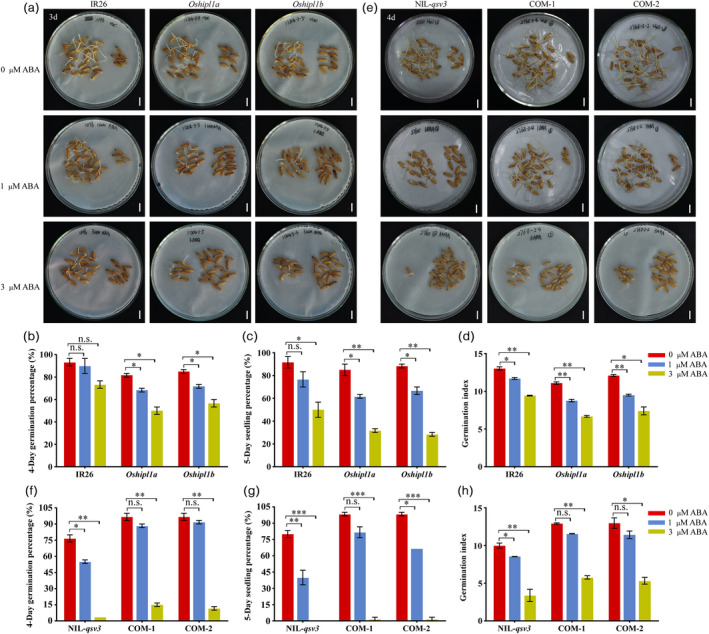
Effects of ABA treatments on OsHIPL1 during seed germination. (a) Photographs of germinated seeds of Oshipl1 mutants and IR26 after 3 days after germination with 0, 1 and 3 μm ABA treatments. (b–d) Comparison of germination percentage (b), seedling percentage (c) and germination index (d) of Oshipl1 mutants and IR26 in the presence of ABA. (e) Photographs of germinated seeds of COM lines and NIL‐qsv3 after 4 days after germination with 0, 1 and 3 μm ABA treatments. (f–h) Comparison of germination percentage (f), seedling percentage (g) and germination index (h) of COM lines and NIL‐qsv3 in the presence of ABA. Bar = 1 cm. Each column presents the means ± standard deviations of three biological replicates. *P < 0.05, **P < 0.01 and ***P < 0.001 compared with the control by Student’s t‐test. n.s. represents no significance.
The Oshipl1 mutants and NIL‐qsv3 line exhibit delayed seed germination, suggesting that the mutants and NIL‐qsv3 line may accumulate more ABA content. We measured the ABA contents among IR26, NIL‐qsv3, Oshipl1a and Oshipl1b lines using an ultra‐performance liquid chromatography–tandem mass spectrometry (LC‐MS/MS) approach during seed germination. The results demonstrated that the ABA levels were significantly increased in germinating seeds of Oshipl1 mutants and NIL‐qsv3 line compared to those in IR26 (Figure 5a), indicating that OsHIPL1 was involved in ABA internal metabolism. Meanwhile, we evaluated the expression levels of pivotal ABA metabolism genes (Figure 5b,c), including OsZEP and OsNCEDs (ABA biosynthesis) and OsABA8oxs (ABA degradation) among IR26, Oshipl1 mutants and NIL‐qsv3 line at 12 h after imbibition. The expression levels of OsZEP and OsNCED4 were significantly up‐regulated in Oshipl1 mutants and NIL‐qsv3 line, compared with IR26, and the expression levels of OsNCED1, OsNCED3 and OsNCED5 were up‐regulated in Oshipl1 mutants while not in NIL‐qsv3 line (Figure 5b). The expression levels of ABA degradation gene OsABA8ox3 were markedly down‐regulated in Oshipl1 mutants and NIL‐qsv3 line, compared with IR26, and the expression levels of OsABA8ox1 were down‐regulated in NIL‐qsv3 line while not in Oshipl1 mutants (Figure 5c). Differential expression levels of OsZEP, OsNCED4 and OsABA8ox3 among IR26, Oshipl1 mutants and NIL‐qsv3 line could well explain the increased ABA content of Oshipl1 mutants and NIL‐qsv3 seeds.
Figure 5.
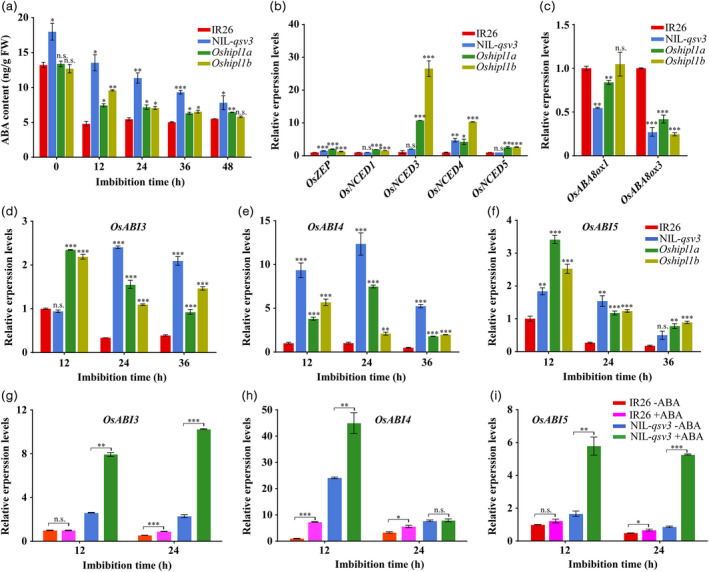
OsHIPL1 altering the contents of ABA and expressions of ABA signalling genes OsABIs during seed germination. (a) The contents of ABA in germinated seeds of IR26, NIL‐qsv3 and Oshipl1 mutants. (b, c) Transcription levels of ABA biosynthesis genes (OsZEP, OsNCED1, OsNCED3, OsNCED4 and OsNCED5) (b) and ABA degradation genes (OsABA8ox1 and OsABA8ox3) (c) among IR26, Oshipl1 mutants and NIL‐qsv3 line at the 12 h seeds after imbibition. The relative expression levels were represented by fold change relative to the expression levels of IR26 at 12h after imbibition. (d–f) Comparison of OsABI3 (d), OsABI4 (e) and OsABI5 (f) expression in germinated seeds among NIL‐qsv3, Oshipl1 mutants and IR26 (control) during seed germination. The relative expression levels were represented by fold change relative to the expression levels of IR26 at 12 h after imbibition. (g–i) Expressions of OsABI3 (g), OsABI4 (h) and OsABI5 (i) in germinated seeds were induced at ABA treatments in IR26 and NIL‐qsv3 using the RT‐qPCR approach. The relative expression levels were represented by fold change relative to the expression levels of IR26 without ABA. The expression of OsHIPL1 was normalized to that of OsActin gene control.
Our global analysis of transcripts in imbibed seeds indicated that the ABA signalling‐related genes, such as OsABI4, OsBZIP23 and OsBZIP12, were significantly induced in NIL‐qsv3 line compared to those of IR26 plants (Figure S8B). We speculate that the regulation of OsHIPL1 on seed vigour might involve in ABA signalling during seed germination in rice. Therefore, we checked the transcript abundance of several identified ABA signalling‐related genes during seed germination, including OsABI3, OsABI4 and OsABI5 (Finkelstein and Lynch, 2000; Kang et al., 2002; Lopez‐Molina et al., 2001; Söderman et al., 2000). The transcript levels of OsABI3, OsABI4 and OsABI5 were significantly increased in NIL‐qsv3 line and Oshipl1 mutants during seed germination compared with those of IR26 (Figure 5d–f). The expression levels of OsABI3, OsABI4 and OsABI5 were further evaluated in the NIL‐qsv3 line and IR26 plants during seed germination under ABA treatment. We observed that the expression levels of OsABI3, OsABI4 and OsABI5 were approximately 2–3 fold higher in NIL‐qsv3 seeds than that without ABA, but weakly induced in IR26 seeds following ABA treatment (Figure 5g–i). These data demonstrate that the disruption of OsHIPL1 alters the expression of OsABI3, OsABI4 and OsABI5, and their continuously higher expressions in germinating seeds caused low seed vigour in Oshipl1 mutants and NIL‐qsv3 line in rice.
OsHIPL1 interacts with aquaporin OsPIP1;1 to improve seed germination
To further understand the molecular mechanism of regulation by OsHIPL1 on seed vigour in rice, we identified the interacting proteins through a yeast two‐hybrid assay with OsHIPL1 as bait. Of the identified 22 candidates (Table S6), 5 candidate proteins were subsequently confirmed by retransformation into yeast, including aquaporin OsPIP1;1 (Figure 6a).
Figure 6.

OsHIPL1 interacts with OsPIP1;1 and promotes water uptake during seed germination. (a) Yeast two‐hybrid screening assay. Interaction of OsHIPL1 with OsPIP1;1 is indicated by the ability of yeast cells to grow on dropout medium lacking Leu, Trp, His and Ade for 4 days after plating. CUB, C‐terminal half of ubiquitin; Nub, N‐terminal half of ubiquitin; NubG, negative control with a point mutation in Nub; NuBI, positive control. The experiments were repeated three times with similar results. (b) Bimolecular fluorescence complementation (BiFC) assay. The fluorescence resulted from the complementation of the C‐terminal portion of YFP fused to OsHIPL1 (OsHIPL1‐cYFP) with the N‐terminal portion of YFP fused to OsPIP1;1 (OsPIP1;1‐nYFP). Fluorescence was observed in tobacco leaf epidermal cells. (c) Firefly luciferase (LUC) complementation imaging assay. cLUC‐OsHIPL1 and nLUC‐OsPIP1;1 with the control vector were co‐infiltrated into N. benthamiana leaves. LUC images were captured at 48 h after infiltration. (d–f) Water contents of Oshipl1 mutants and IR26 (d), COM lines and NIL‐qsv3 (e) and OE lines and ZH11 (f) during seed germination. (g) Comparison of the transcription levels of OsPIP1;1 among IR26, Oshipl1 mutants, NIL‐qsv3, COM lines, ZH11 and OE lines at 24 h after imbibition using the RT‐qPCR approach. The relative expression levels were represented by fold change relative to the expression levels of IR26 or ZH11. The expression of OsPIP1;1 was normalized to that of OsActin gene control. (h) Transcription levels of OsPIP1;1 response to ABA treatments in germinated seeds were conducted using the RT‐qPCR approach. The relative expression levels were represented by fold change relative to the expression level of OsPIP1;1 at 12 h after imbibition without ABA. Each column presents the means ± standard deviations of three biological replicates. *P < 0.05, **P < 0.01 and ***P < 0.001 compared with the control by Student’s t‐test. n.s. represents no significance.
To further confirm the OsHIPL1‐OsPIP1;1 interaction, the bimolecular fluorescence complementation (BiFC) and luciferase (LUC) assays were conducted. The yellow fluorescent protein (YFP) signals were only observed on the plasma membrane of N. benthamiana leaves when cYFP‐OsHIPL1 was co‐infiltrated with nYFP‐OsPIP1;1 (Figure 6b). Meanwhile, only co‐expression of cLUC‐OsHIPL1 and nLUC‐OsPIP1;1 in tobacco leaves could reconstitute LUC activity compared with the various negative controls (Figure 6c). These results demonstrate that OsHIPL1 could interact with OsPIP1;1.
To investigate whether OsHIPL1 affects water uptake during seed germination, we tested the water content of the germinating seeds. The water content of Oshipl1 mutants was significantly reduced compared with that of IR26 (Figure 6d). By contrast, the water content of COM and OE lines was significantly increased compared with that of NIL‐qsv3 line and ZH11, respectively (Figure 6e,f). These data indicate that OsHIPL1 promotes uptake of water during seed germination.
The OsPIP1;1 expression was also tested. A reduction in the transcript levels of OsPIP1;1 in Oshipl1 mutants was observed compared with that in IR26, and an increase in the relative expression levels of OsPIP1;1 in COM lines was observed compared with that in NIL‐qsv3 line (Figure 6g). There were no obvious differences in the expression levels of OsPIP1;1 of OE lines compared with that in wild‐type ZH11.
It was well‐known that ABA could regulate the expression of aquaporin genes in a variety of plants (Jang et al., 2004; Mariaux et al., 1998; Suga et al., 2002; Zhu et al., 2005). Therefore, the transcript levels of OsPIP1;1 were tested during seed germination at the treatments with ABA. The results showed that the expression of OsPIP1;1 was significantly reduced during seed germination in the presence of ABA (Figure 6h), suggesting ABA may inhibits OsPIP1;1 expression.
Genetic variation in the regulatory region of OsHIPL1
To identify the allelic constitution of OsHIPL1, 192 rice accessions of the Rice Diversity Panel 1 (RDP1) were selected to be evaluated by GP, SP and GI during seed germination (Table S7). Four SNPs were identified in the coding region of OsHIPL1 and three haplotypes (Hap1, Hap2 and Hap3) were detected (Figure 7a). The 4‐ and 5‐day GP, 6‐ and 7‐day SP of Hap1 were significantly higher than those of Hap2 and Hap3 (Figure 7b–f), suggesting Hap1 is superior allele for seed vigour. These four SNPs, SNP‐3.27670307., SNP‐3.27672138., SNP‐3.27673895. and SNP‐3.27674048., were consistent with SNP2, SNP3, SNP5 and SNP7 identified between IR26 and Jiucaiqing (Table S7) respectively. Of them, the amino acid residue change in SNP3 is from serine to asparagine (Ser1415Asn) in GSDH domain (Figure 1d,e), suggesting that SNP3 may be significantly associated with seed vigour. To confirm this hypothesis, we further analysed the seed vigour of 192 rice accessions. The results showed that 126 rice accessions harbouring ‘A’ SNP exhibited higher GP and SP than did 66 rice accessions with ‘G’ SNP (Figure 7g–k). These results indicated that Hap1 and SNP3 of qSV3 have significantly positive correlation with seed vigour in rice.
Figure 7.
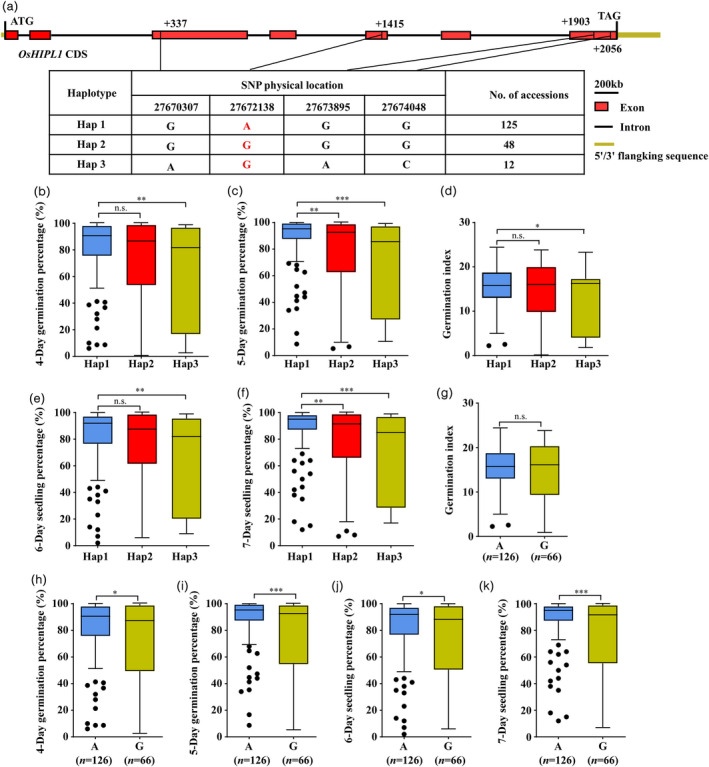
Haplotypes of OsHIPL1 associated with seed vigour in rice. (a) Haplotypes of OsHIPL1 were identified in the full‐length CDS of the gene. Red boxes represent the exon; solid lines represent the intron; yellow boxes represent 5’UTR and 3’UTR. (b–f) Comparison of the germination percentage, seedling percentage and germination index between accessions harbouring different haplotypes. (g–k) Comparison of the germination percentage, seedling percentage and germination index of accessions harbouring 1415A or 1415G SNP. The number of rice accessions is listed in brackets. (*P < 0.05; **P < 0.01, ***P < 0.001 compared with the control by Student’s t‐test. n.s. represents no significance).
OsHIPL1 increases the seed vigour of direct seeding rice under greenhouse and field
To investigate the application of OsHIPL1 for direct seeding, seedling establishment and seedling growth were compared among IR26, Oshipl1 mutants, NIL‐qsv3, the COM lines and ZH11 and OE lines when seeds were directly sown in soil. In the greenhouse, the SP, root length, shoot length and basal shoot diameter were significantly higher in IR26 than in NIL‐qsv3 line and Oshipl1 mutants (Figure 8). Moreover, the COM lines exhibited a significant increase in the SP, shoot length, shoot length and basal shoot diameter compared with those of NIL‐qsv3 line, and the OE lines significantly increase in the shoot length and basal shoot diameter compared with those of ZH11 (Figure 8). Meanwhile, there were the decreases in the SP of NIL‐qsv3 line and Oshipl1 mutants compared with those of IR26 and the increases in the SP of COM lines compared with those of NIL‐qsv3 line under the field (Figure S10). Agronomic traits were similar between IR26, Oshipl1 mutants, NIL‐qsv3 and COM lines, including plant height, effective tillers, 1000‐grain weight, panicle length, setting percentage and grain per panicle (Figure S11). These results suggested that qSV3 could improve the performance of plants directly seeded without adverse influence on other agronomic traits.
Figure 8.

Effects of OsHIPL1 on seedling growth under direct seeding in soil. (a–c) Photographs of seedling establishment of Oshipl1 mutants and IR26 (a), COM lines and NIL‐qsv3 (b), and OE lines and ZH11 (c) at 5 days after sowing. (d–f) Photographs of seedling growth of Oshipl1 mutants and IR26 (d), COM lines and NIL‐qsv3 (e) and OE lines and ZH11 (f) at 10 days after sowing. (g–i) Photographs of representative seedlings of Oshipl1 mutants and IR26 (g), COM lines and NIL‐qsv3 (h) and OE lines and ZH11 (i) at 10 days after sowing. (j–m) Comparison of seedling percentage (j), root length (k), shoot length (l) and base diameter (m) among IR26, Oshipl1 mutants, NIL‐qsv3, COM lines, ZH11 and OE lines. Bars = 1 cm. Each column presents the means ± standard deviations of three biological replicates. *P < 0.05, **P < 0.01 and ***P < 0.001 compared with the control by Student's t‐test. n.s. represents no significance.
Discussion
Seed vigour is an important trait for production of direct seeding of rice, which could be evaluated by the speed and uniformity of seed germination, and seedling establishment (Bewley et al., 2013; Rajjou et al., 2012). In this study, seven QTLs were identified, related to GP, SP and GI of rice seed vigour, using one CSSL population under H2O condition respectively. Three major QTLs qGP3.2, qGI3.2 and qSP3.1 were integrated into a qSV3 locus, and further confirmed by the segregation population derived from the residual heterozygous CSSL10 line. Compared with the reported QTLs, the qSV10 (qGP10.1 and qSP10.1) is a novel QTL for seed vigour; the qSV3 is located in the downstream of OsHAK21 (He et al., 2019), co‐localized with qGR3.3 identified under H2O condition (Zeng et al., 2021) and qLTG‐3‐2 under low‐temperature stress (Fujino et al., 2004). As the seed vigour of Oshipl1 mutants and NIL‐qsv3 was significantly reduced under abiotic stresses, and the seed vigour of COM lines was restored (Figures S12 and S13), it implied that qSV3 could also respond to abiotic stresses, with similar function of qLTG‐3‐2 reported (Fujino et al., 2004). Its molecular regulation mechanism and application in improvement of seed vigour in rice breeding deserve to be further investigated.
We isolated and characterized the causal gene OsHIPL1 of qSV3 in this study, which encodes a hedgehog‐interacting protein‐like 1 protein. Human HHIPL1 was shown to positively regulate hedgehog signalling (Aravani et al., 2019) and the hedgehog signalling in mammalian was reported to modulate the embryogenesis, cancer and lifespan (Katoh and Katoh, 2006; Arraf et al., 2020; Rallis et al., 2020; Jiang, 2021). A phylogenetic tree analysis shows that HIPL1 protein is highly conserved in plants, such as rice, Brachyodium stacei, maize, sorghum and Arabidopsis (Figure S14). Moreover, the function of HIPL1 protein is still unknown in plants. Protein structure analysis showed that OsHIPL1 contained one transmembrane domain, three low complexity regions and one GSDH domain. Five SNPs in IR26 caused amino acid residue change in the coding region compared with Jiucaiqing, including SNP3 and SNP6 located in GSDH domain and the third low complexity region respectively. This change may result in higher seed vigour of IR26 than Jiucaiqing.
It is well‐known that ABA is a sesquiterpenoid hormone that plays central roles in seed maturation, germination and stress responsiveness (Finkelstein et al., 2002). In our study, the ABA content in germinating seeds of Oshipl1 mutants and NIL‐qsv3 line was significantly higher than that of IR26 plants (Figure 5a), indicating that the decrease in seed vigour of Oshipl1 mutants and NIL‐qsv3 line might be due to the increased level of endogenous ABA in seeds. We hypothesized that the seed germination controlled by OsHIPL1 is involved in ABA metabolism. It is also confirmed by the reduced transcript abundance of OsABA8ox3 and increased transcript levels of the OsZEP and OsNCED4 in NIL‐qsv3 line and Oshipl1 mutants compared with IR26 (Figure 5b,c). Transcription factors, ABI3, ABI4 and ABI5, as positive regulators in ABA signalling, contributed to delayed seed germination in plants (Albertos et al., 2015; Nambara et al., 1995; Nonogaki, 2019; Shu et al., 2013; Zhang et al., 2005). The higher transcript levels of OsABI3, OsABI4 and OsABI5 were observed in germinating seeds of NIL‐qsv3 line and Oshipl1 mutants compared with IR26 in this study (Figure 5d–f). It indicates that OsHIPL1 modulated the expression of OsABI3, OsABI4 and OsABI5. Previous studies show that increased expression of OsABI3, OsABI4 and OsABI5 reduces seed germination and seedling establishment (He et al., 2020; Nonogaki, 2019). These results indicate that the higher endogenous ABA and higher expressions of OsABIs in germinating seeds of Oshipl1 mutants and NIL‐qsv3 line lead to the decreases in seed vigour in mutants and NIL‐qsv3 line. We also measured IAA content of oshipl1a, oshipl1b and IR26 in germinated seeds, while no obvious differences were identified between oshipl1 mutants and IR26 (Figure S15). It hints that function of OsHIPL1 on seed vigour might be different from that of OsIAGLU, which regulates seed vigour through mediating crosstalk between IAA and ABA reported by He et al. (2020).
Aquaporins, belonging to the highly conserved major intrinsic protein (MIP) family, play an essential role in plant water transport (Footitt et al., 2019; Liu et al., 2007, 2013; Roche and Törnroth‐Horsefield, 2017). The co‐expression of aquaporins ZmPIP1;2 and ZmPIP2;1 in X. laevisoo cytes increased the cell osmotic water permeability coefficient (P f) (Fetter et al., 2004). Zelazny et al. (2007) demonstrated that interactions between ZmPIP1s and ZmPIP2s occurred in maize mesophyll protoplasts, and the PIP1–PIP2 interaction induced the relocation of ZmPIP1s to the plasma membrane. Liu et al. (2013) reported that co‐expression of OsPIP2;1 with OsPIP1;1 could help OsPIP1;1 incorporating into plasma membrane and significantly increasing its water permeability. Our results showed that OsHIPL1 could interact with OsPIP1;1 and co‐locate in the plasma membrane. It suggested that OsHIPL1, like OsPIP2;1, might make OsPIP1;1 incorporating into plasma membrane and significantly increase water uptake for seed germination. The molecular mechanism of OsHIPL1 interacting with OsPIP1;1 warrants further investigation in the future.
As previously reported, the moderate expression of OsPIP1;1 increased seed germination of rice (Liu et al., 2013). Similar results were observed in this stud;, the alteration of expression levels of OsPIP1;1 was observed in Oshipl1 mutants, NIL‐qsv3 and COM lines during seed germination, while not in OE lines (Figure 6g). It indicates that the excess OsHIPL1 proteins could not enhance the expression levels of OsPIP1;1 during seed germination. The expression of OsHIPL1 was increased under ABA treatments during seed germination (Figure S9), while the expression of OsPIP1;1 was decreased (Figure 6h). We proposed that under normal condition (without ABA treatment), OsHIPL1 could up‐regulate OsPIP1;1 expression, but under ABA treatment, the excess expression of OsHIPL1 induced could not synchronously affect the OsPIP1;1 expression. Guo et al. (2006) reported that the OsPIP1;1 expression was significantly inhibited by ABA treatment in roots in the seedling stage of rice, similar to our results.
ABA can influence activity of aquaporins at different levels by either changing the expression of aquaporin genes or post‐transcriptional modifications (Jang et al., 2004; Mariaux et al., 1998; Suga et al., 2002; Zhu et al., 2005). ABA‐induced dephosphorylation of aquaporins leads to reduced water flux (Kline et al., 2010). ABA‐dependent changes in dormancy and germination also result from shifting water potential thresholds for radicle emergence (Ni and Bradford, 1992). In Arabidopsis, the expression of CYP707A2, which encodes an ABA 8′‐hydroxylase, rapidly increases after seed imbibition and plays a key role in inactivating ABA during germination (Kushiro et al., 2004). We thus speculated that the crosstalk of ABA metabolism and seed imbibition might influence seed vigour. In our study, the Oshipl1 mutants have lower imbibition rate that might cause lower ABA degradation in germinating seeds, but the molecular mechanism needs to be further investigated.
Summary, we clarified the function of hedgehog‐interacting protein‐like 1 protein gene OsHIPL1 in plants. Although it is unknown whether OsHIPL1 has similar pathway as hedgehog signalling in plants as in humans, there are two possible pathways by which OsHIPL1 regulates seed vigour in rice (Figure 9). On the one hand, OsHIPL1 contributed to seed germination and seedling establishment involved in modulating ABA levels and ABA signalling in germinating seeds in rice. On the other hand, OsHIPL1 interacted with the OsPIP1;1, and regulated the water uptake to improve rice seed vigour. However, it is worthy to further study how OsHIPL1 influences the expressions of OsABIs, and what other factors in the interaction of OsHIPL1 and OsPIP1;1 regulate the germination of seeds. Our studies provide a novel gene OsHIPL1 and its information on the molecular mechanisms of seed vigour. Meanwhile, the superior haplotype Hap1 and its linkage molecular marker SNP3 of OsHIPL1 were identified, which would facilitate improvement in seed vigour in rice breeding programme and be beneficial to production of rice direct seeding.
Figure 9.

Proposed model for the role of OsHIPL1 in seed vigour in rice. This model shows that OsHIPL1 might reduce the endogenous ABA contents in germinating seeds, which could decrease the expression levels of the ABA signalling‐related genes ABI3/4/5, and then improve rice seed vigour. In addition, OsHIPL1 might affect the expression of OsPIP1;1, and the complex of OsHIPL1 with OsPIP1;1 could regulate water uptake to increase seed vigour.
Materials and methods
Plant materials and growth conditions
Japonica Jiucaiqing and indica IR26, and 62 CSSLs developed by introgressing chromosome segments of Jiucaiqing into IR26 (Cheng et al., 2016, Tables S1 and S2), were used for QTL mapping. A residual heterozygous line CSSL10 (BC6F2) and its selfing population were used for the confirmation of the major qSV3. One heterozygous line (BC6F3) between markers HY27 and HY44 from the selfing segregation population of line CSSL10 was selected to develop the genetic population I (BC6F4, 5000 individuals) and population II (BC6F5, 992 individuals) by selfing for fine mapping of the qSV3 locus. A homozygous line containing the allele (qsv3) from Jiucaiqing between markers HY42 and HY43 was selected for the development of NIL‐qsv3. In total, 192 accessions from the rice diversity panel (Eizenga et al., 2014) were used for haplotype analyses of qSV3 locus (Table S7). All plants were grown in an experimental field at Nanjing Agricultural University, Nanjing, Jiangsu. All seeds were harvested at their maturity stage and dried at 42 °C for 7 days to break seed dormancy.
Evaluation of seed vigour
Seed germination was conducted as previously described by Wang et al. (2010). Thirty seeds of each genotype were imbibed in Petri dishes (diameter 9 cm) with 10 mL distilled water at 30 ± 1 °C for 7 days. Seeds were also treated with 10 mL of 20% PEG6000 (polyethylene glycol with an average molecular weight of 6000 Da), 300 mm mannitol and 20 mL of 200 mm NaCl at 30 ± 1 °C for 11 days, respectively, and with 10 mL of distilled water at 15 ± 1 °C for 11 days. As to ABA treatments, distilled water was replaced with the solution containing 0, 1 and 3 μm of ABA respectively. The number of germinated seeds was observed daily. Germination was defined as the emergence of the radicle through the hull by ≥2 mm. Seedlings were considered to have developed when the root length reached the seed length and the shoot length reached half of the seed length (Xu et al., 2017). Three replications were performed. GP, SP and GI were calculated as follows: GP = (total germinated seeds/30) × 100%, SP = (total seedlings/30) × 100% and GI = ∑(Gt/t), where Gt is the number of germinated seeds on Day t (Wang et al., 2010).
Map‐based cloning
The GP, GI and SP of the individual among CSSLs were used by SMA for QTL mapping, and the confirmation of major qSV3 was conducted by ICIM (Li et al., 2008). Genomic DNA of seedlings from the population I and population II was extracted using the cetyltrimethylammonium bromide (CTAB) method to screen recombinants for fine mapping (Murray and Thompson, 1980). Markers used for QTL mapping are listed in Table S7. The candidate gene was predicted using the data at http://rice.plantbiology.msu.edu/cgi‐bin/gbrowse. The cDNA sequences of the predicted gene were amplified from Jiucaiqing and IR26 using Phanta Super‐Fidelity DNA polymerase (Vazyme Biotech Co., Ltd, Nanjing, Jiangsu, China).
Plasmid construction and plant transformation
For generating OsHIPL1‐overexpressing lines, the full‐length coding sequence of OsHIPL1 was cloned into PBWA(V)HS, driven by 35S promoter and then transformed into japonica Zhonghua11 (ZH11) by Agrobacterium tumefaciens‐mediated transformation (Hiei et al., 1994). The Oshipl1 mutants (IR26 background) were generated by CRISPR‐Cas9 system (Xing et al., 2014). To produce the complementation construct PBWA(V)HS‐OsHIPL1 Pro‐c OsHIPL1, the full‐length CDS of OsHIPL1 (2079 bp) was cloned by RT‐qPCR amplification. The CDS containing the entire OsHIPL1 coding region was inserted into the modified vector PBWA(V)HS, and then the OsPHF1 promoter was amplified from IR26 genomics DNA and inserted before OsHIPL1‐coding region to generate the vector PBWA(V)HS‐OsHIPL1 Pro‐c OsHIPL1. The vectors were introduced into NIL‐qsv3 line using the Agrobacterium‐mediated transformation method. All DNA constructs were confirmed by sequencing, and all transgenic plants were identified by the RT‐qPCR and hygromycin gene amplification. Specific primers were designed to confirm the mutation positions in each CRISPR/Cas9‐positive transgenic line. All primers are listed in Table S8.
Expression analysis
Total RNA was extracted from the developing grains (0, 7, 14, 28 and 35 days after flowering; DAF), germinating seeds (0, 6, 12, 24, 30, 36, 48, 54, 60 and 72 h imbibition) and different tissues (root, stem, leaf, sheath, node and spike) of IR26 using the HP Plant RNA kit (Omega, Atlanta), according to the manufacturer's instructions. First‐strand cDNA was synthesized with random oligonucleotides using the HiScriptII Reverse Transcriptase system (Vazyme Biotech Co., Ltd.). The RT‐qPCR was performed on a Roche Light Cycler 480 system using SYBR Green Mix. The Actin gene was used as the internal control for normalization. Primers used for expression analysis are listed in Table S8. Three biological replicates were made.
Subcellular localization
The open reading frame of OsHIPL1 (without the stop codon) was amplified and inserted into the pCambia1300s‐GFP vector driven by the CaMV 35S promoter and the PAN580‐GFP vector driven by the CaMV 35S promoter according to the manufacturer’s instructions (Vazyme, Nanjing, China). Then, the construct was introduced into Agrobacterium tumefaciens strain EH105 and infiltrated into rice protoplasts and N. benthamiana leaves (Chen et al., 2008). Fluorescence signals were observed using a LSM 710 confocal microscope (Zeiss, Oberkochen, Germany).
Phylogenetic analysis
The entire amino acid sequences of OsHIPL1 were used as the query to search for the homologous proteins in the NCBI website (https://blast.ncbi.nlm.nih.gov/Blast.cgi) and GBI website (https://phytozome.jgi.doe.gov). All HIPL1 proteins were clustered using ClustalX, and the phylogenetic tree was generated by MEGA6 based on the neighbour‐joining method and bootstrap analysis (1000 replicates).
Hormone quantification
The extraction and quantification of endogenous ABA were conducted according to the manufacturer’s instructions (Wuhan Metware Biotechnology Co., Ltd., Wuhan, China). The ABA content was quantified by LC‐MS/MS system as described by He et al. (2020). Three biological replicates were conducted.
RNA‐seq analysis
The IR26 and NIL‐qsv3 seeds were grown on media for 12 h, and then the materials were collected. Three biological replicates were prepared for each sample. Total RNA was extracted using HP Plant RNA kit (Omega, Atlanta, GA) according to the manufacturer’s instructions and then sequenced (PERSONAL, Nanjing, China). Clean reads were mapped to the japonica rice Nipponbare reference genome after screening and trimming. Genes with P‐value < 0.05 and fold change ≥2 were identified as reliable differentially expressed genes (DEGs). Multiple testing was corrected via false discovery rate estimation and q‐values below 0.05 were considered to indicate differential expression. Twelve genes were randomly selected for the confirmation of RNA‐seq by the RT‐qPCR and their primers are listed in Table S8.
Y2H screening and confirmation
DUAL Membrane Pairwise Interaction kit (Dualsystems Biotech) and cDNA library of seeds were used for the Y2H assays. The full‐length CDS of OsHIPL1 was fused to PBT3‐N to construct the bait vector, which was transformed into the yeast strain NMY51. Yeast screening was performed as described in the manufacturer's protocol (Clontech Yeast Protocols Handbook). To confirm protein–protein interaction in yeast, the full‐length CDS of OsPIP1;1 was cloned into the pPR3‐N vector to generate NubG‐OsPIP1;1. The recombinant plasmid pairs were co‐transformed into the yeast strain NMY51. The transfected yeast cells were plated on SD/‐Leu/‐Trp medium and SD/‐Ade/‐His/‐Leu/‐Trp medium and cultured at 28 °C for 4 days. The primers and restriction enzyme sites were used to amplify sequences and generate vectors. The relevant primers are listed in Table S8.
BiFC assay
The OsHIPL1 and OsPIP1;1 cDNA fragments were cloned into p2YN and p2YC to form the nYFP protein and cYFP protein constructs respectively. The plasmids were then transformed into A. tumefaciens strain EH105 and all constructs were verified by DNA sequencing. For the transient expression assay, these constructs were co‐infiltrated with the p19 strain into 5‐week‐old N. benthamiana leaves. YFP fluorescence was excited at 515 nm by a Zeiss LSM780 confocal scanning microscope in dark for 48 hours after infiltration. Primers used to generate the constructs are listed in Table S8.
Luciferase (LUC) assays
The pCAMBIA1300‐cLUC‐OsHIPL1 and pCAMBIA1300‐nLUC‐OsPIP1;1 plasmids were generated by CDS of OsHIPL1 and OsPIP1;1. The LUC assays were performed as previously described with some modifications (Chen et al., 2008). Different recombinant plasmids including nLUC‐OsPIP1;1 and cLUC‐OsHIPL1 with the control vector were introduced into Agrobacterium strain EH105. Agrobacteria cultures were resuspended overnight with infiltration buffer (10 mm MgCl2, 0.1 mm acetosyringone and 10 mm MES). Different experiment and control group agrobacteria suspension were mixed and co‐infiltrated into 5‐ to 6‐week‐old Nicotiana benthamiana leaves by using a needleless syringe, then weak light growth. Luciferin (1 mm) was sprayed onto the leaves, and the plants were kept in the dark for 2–5 min. LUC images were captured using a cooled CCD imaging apparatus (Chen et al., 2008). The primers were listed in Table S8.
Haplotype analysis
To determine the haplotypes of the OsHIPL1, the 700 000 SNP markers of rice accessions at https://ricediversity.org/data/index.cfm (McCouch et al., 2016) were used. Four SNPs in the full‐length coding region of OsHIPL1 were located according to the Rice Genome Annotation Project MSU7 database (Rice Genome Browser: http://rice.plantbiology.msu.edu). Seed germination was conducted using 192 accessions under H2O condition for 7 days (Table S7). The haplotypes represented at least 10 investigated accessions that were previously used for a comparative analysis of phenotype (Dong et al., 2016).
Direct sowing rice assay
Forty healthy seeds of IR26, NIL‐qsv3, ZH11, Oshipl1 mutants, complementation and overexpression lines were sowed 1 cm deep in soil in the same plot in an incubator with a 14 h : 10 h, light : dark cycle at 30 °C : 25 °C with 100% relative humidity. Similarly, wild‐type and transgenic lines were sowed 1 cm deep in rice fields with water saturation of 100% for 22 days in June 2021, during which the average high and low temperature ranged from 22 to 29 °C. The SP on the 5th day, root length, shoot length and basal shoot diameter on the 10th day were investigated and calculated. Three biological replicates were made.
Data analysis
Experimental data were analysed using the SAS software (Cary, NC), and significant differences among samples were compared using Student’s t‐test or fisher’s least significant difference (LSD) test at the 5%, 1% and 0.1% levels of probability.
Conflicts of interest
The authors declare no conflict of interest.
Author contributions
Ying He, Jinping Cheng and Hongsheng Zhang planned the research, analysed the data and drafted the manuscript. Ying He performed all important experiments. Shanshan Chen, Kexin Liu, Yongji Chen, Yanhao Cheng, Peng zeng, Peiwen Zhu and Ting Xie performed part of the seed germination experiments. Sunlu Chen provided reliable experimental advice.
Funding
This work was supported by the Hainan Yazhou Bay Seed Laboratory (Project of B21HJ1002), the Natural Science Foundation of Jiangsu Province (Grant No. SBK2020020068) and the National Natural Science Foundation of China (Grant No. 32172037, 32000377 and 31771757).
Supporting information
Figure S1 Comparison of seed germination and seedling establishment between JCQ and IR26 under normal condition.
Figure S2 QTLs analysis of seed germination traits among F2 population under normal condition.
Figure S3 Comparison of coding sequences in ORF2 between IR26 and Jiucaiqing.
Figure S4 Comparison of seed vigour between NIL‐qsv3 and IR26 under normal condition.
Figure S5 Comparisons of the OsHIPL1 transcript levels between transgenic lines (OE1, OE2, COM‐1 and COM‐2) and wild type (NIL‐qsv3 and ZH11).
Figure S6 The confirmation of differential expression genes (DEGs) between IR26 and NIL‐qsv3 in 12h imbibed seeds via the RT‐qPCR approach in rice.
Figure S7 GO enrichment analysis for differentially expressed genes (DEGs) in IR26 compared to NIL‐qsv3.
Figure S8 Rice OsHIPL1 altering expressions of genes involved in ABA signalling during seed germination.
Figure S9 Transcription levels of OsHIPL1 response to ABA treatments in germinating seeds conducted using the RT‐qPCR approach.
Figure S10 Comparison of seedling establishment and seedling growth between wild‐type and the transgenic lines when seeds are directly sown in outdoor field soil.
Figure S11 Comparison of agronomic traits among IR26, Oshipl1 mutants, NIL‐qsv3 and COM lines.
Figure S12 Comparison of seed germination percentage and seedling percentage between NIL‐qsv3 and IR26 under low temperature (A–C), salinity (D–F), PEG (G–I) and mannitol (J–L) stresses.
Figure S13 Comparison of seed germination percentage and seedling percentage of the transgenic lines of OsHIPL1 under salinity (A‐D) and PEG (E‐H) stresses.
Figure S14 Phylogenetic relationships of OsHIPL1 among Oryza sativa, Maize, Sorghum, Setaria italica, Setaria viridis, Brachypodium, Brachypodium stacei, Arabidopsis, Prunus persica and Glycine max.
Figure S15 The contents of IAA in germinated seeds of Oshipl1 mutants and IR26.
Table S1 The information of phenotype of seed vigour of the CSSLs and parents.
Table S2 The information of genotype of seed vigour of the CSSLs and parents.
Table S3 The information QTLs of seed vigour identified by single marker mapping based on the CSSL population.
Table S4 Differential expression genes (DEGs) between IR26 and NIL‐qsv3 in 12h imbibed seeds in rice.
Table S5 DEGs involved in hormone metabolism between IR26 and NIL‐qsv3 in 12h imbibed seeds in rice.
Table S6 Identification of putative OsHIPL1‐interacting proteins using yeast two‐hybrid screening.
Table S7 Information of the 192 accessions used for haplotype analyses in rice.
Table S8 The primer pairs used in this study.
Acknowledgements
We thank Dr. Jian Hua at Cornell University and Nanjing Agricultural University for seeds and Dr. Susan McCouch at Cornell University for seeds of the Rice Diversity Panel.
He, Y. , Chen, S. , Liu, K. , Chen, Y. , Cheng, Y. , Zeng, P. , Zhu, P. , Xie, T. , Chen, S. , Zhang, H. and Cheng, J. (2022) OsHIPL1, a hedgehog‐interacting protein‐like 1 protein, increases seed vigour in rice. Plant Biotechnol. J., 10.1111/pbi.13812
Contributor Information
Hongsheng Zhang, Email: hszhang@njau.edu.cn.
Jinping Cheng, Email: cjp@njau.edu.cn.
References
- Abe, A. , Takagi, H. , Fujibe, T. , Aya, K. , Kojima, M. , Sakakibara, H. , Uemura, A. et al. (2012) OsGA20ox1, a candidate gene for a major QTL controlling seedling vigor in rice. Theor. Appl. Genet. 125, 647–657. [DOI] [PubMed] [Google Scholar]
- Albertos, P. , Romero‐Puertas, M.C. , Tatematsu, K. , Mateos, I. , Sanchez‐Vicente, I. , Nambara, E. and Lorenzo, O. (2015) S‐nitrosylation triggers ABI5 degradation to promote seed germination and seedling growth. Nat. Commun. 6, 8669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravani, D. , Morris, G.E. , Jones, P.D. , Tattersall, H.K. , Karamanavi, E. , Kaiser, M.A. , Kostogrys, R.B. et al. (2019) HHIPL1, a gene at the 14q32 coronary artery disease locus, positively regulates hedgehog signaling and promotes atherosclerosis. Circulation, 140, 500–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arraf, A.A. , Yelin, R. , Reshef, I. , Jadon, J. , Abboud, M. , Zaher, M. , Schneider, J. et al. (2020) Hedgehog signaling regulates epithelial morphogenesis to position the ventral embryonic midline. Dev. Cell, 53, 589–602. [DOI] [PubMed] [Google Scholar]
- Bailly, C. , El‐Maarouf‐Bouteau, H. and Corbineau, F. (2008) From intracellular signaling networks to cell death: the dual role of reactive oxygen species in seed physiology. C R Biol. 331, 806–814. [DOI] [PubMed] [Google Scholar]
- Bewley, J.D. (1997) Seed germination and dormancy. Plant Cell, 9, 1055–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley, J.D. , Bradford, K. , Hilhorst, H. and Nonogaki, H. (2013) Seeds: Physiology of Development, Germination and Dormancy, 3rd ed. Heidelberg: Springer. [Google Scholar]
- Borner, G.H. , Lilley, K.S. , Stevens, T.J. and Dupree, P. (2003) Identification of glycosylphosphatidylinositol‐anchored proteins in Arabidopsis. A proteomic and genomic analysis. Plant Physiol. 132, 568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner, G.H. , Sherrier, D.J. , Stevens, T.J. , Arkin, I.T. and Dupree, P. (2002) Prediction of glycosylphosphatidylinositol‐anchored proteins in Arabidopsis. A genomic analysis. Plant Physiol. 129, 486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H.M. , Zou, Y. , Shang, Y.L. , Lin, H.Q. , Wang, Y.J. , Cai, R. , Tang, X.Y. et al. (2008) Firefly luciferase complementation imaging assay for protein‐protein interactions in plants. Plant Physiol. 146, 368–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, K. , Li, G.J. , Bressan, R.A. , Song, C.P. , Zhu, J.K. and Zhao, Y. (2020) Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 62, 25–54. [DOI] [PubMed] [Google Scholar]
- Cheng, J.P. , He, Y.Q. , Zhan, C.F. , Yang, B. , Xu, E.S. , Zhang, H.S. and Wang, Z.F. (2016) Identification and characterization of quantitative trait loci for shattering in rice Landrace Jiucaiqing from Taihu Lake Valley, China. Plant Genome, 9, 1–9. [DOI] [PubMed] [Google Scholar]
- Chuang, P.T. , Kawcak, T. and McMahon, A.P. (2003) Feedback control of mammalian Hedgehog signaling by the Hedgehog‐binding protein, Hip1, modulates Fgf signaling during branching morphogenesis of the lung. Genes Dev. 17, 342–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuang, P.T. and McMahon, A.P. (1999) Vertebrate Hedgehog signalling modulated by induction of a Hedgehog‐binding protein. Nature, 397, 617–621. [DOI] [PubMed] [Google Scholar]
- Cutler, S.R. , Rodriguez, P.L. , Finkelstein, R.R. and Abrams, S.R. (2010) Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. [DOI] [PubMed] [Google Scholar]
- Debeaujon, I. and Koornneef, M. (2000) Gibberellin requirement for Arabidopsis seed germination is determined both by testa characteristics and embryonic abscisic acid. Plant Physiol. 122, 415–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding, Z.J. , Yan, J.Y. , Li, G.X. , Wu, Z.C. , Zhang, S.Q. and Zheng, S.J. (2014) WRKY41 controls Arabidopsis seed dormancy via direct regulation of ABI3 transcript levels not downstream of ABA. Plant J. 79, 810–823. [DOI] [PubMed] [Google Scholar]
- Dong, H. , Zhao, H. , Xie, W. , Han, Z. , Li, G. , Yao, W. , Bai, X. et al. (2016) A novel tiller angle gene, TAC3, together with TAC1 and D2 largely determine the natural variation of tiller angle in rice cultivars. PLoS Genet. 12, e1006412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiji, N. , Masanori, O. , Kiyoshi, T. , Ryoichi, Y. , Mitsunori, S. and Yuji, K. (2010) Abscisic acid and the control of seed dormancy and germination. Seed Sci. Res. 20, 55–67. [Google Scholar]
- Eizenga, G.C. , Ali, M.L. , Bryant, R.J. , Yeater, K.M. , McClung, A.M. and McCouch, S.R. (2014) Registration of the Rice Diversity Panel 1 for genome wide association studies. J. Plant Reg. 8, 109–116. [Google Scholar]
- El‐Maarouf‐Bouteau, H. and Bailly, C. (2008) Oxidative signaling in seed germination and dormancy. Plant Signal Behav. 3, 175–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetter, K. , Van Wilder, V. , Moshelion, M. and Chaumont, F. (2004) Interactions between plasma membrane aquaporins modulate their water channel activity. Plant Cell, 16, 215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R. , Gampala, S.S. and Rock, C.D. (2002) Abscisic acid signaling in seeds and seedlings. Plant Cell, 14(Suppl), S15–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R.R. and Lynch, T.J. (2000) The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell, 12, 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein, R. , Reeves, W. , Ariizumi, T. and Steber, C. (2008) Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59, 387–415. [DOI] [PubMed] [Google Scholar]
- Foolad, M.R. , Subbiah, P. and Zhang, L. (2007) Common QTL affect the rate of tomato seed germination under different stress and non stress conditions. Int. J. Plant Genomics, 2007, 97386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt, S. , Clewes, R. , Feeney, M. , Finch‐Savage, W.E. and Frigerio, L. (2019) Aquaporins influence seed dormancy and germination in response to stress. Plant Cell Environ. 42, 2325–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujino, K. , Sekiguchi, H. , Sato, T. , Kiuchi, H. , Nonoue, Y. , Takeuchi, Y. , Ando, T. et al. (2004) Mapping of quantitative trait loci controlling low‐temperature germinability in rice (Oryza sativa L.). Theor. Appl. Genet. 108, 794–799. [DOI] [PubMed] [Google Scholar]
- Fujino, K. , Sekiguchi, H. , Matsuda, Y. , Sugimoto, K. , Ono, K. and Yano, M. (2008) Molecular identification of a major quantitative trait locus, qLTG3‐1, controlling low‐temperature germinability in rice. Proc. Natl Acad. Sci. USA, 105, 12623–12628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommers, C.M.M. and Monte, E. (2018) Seedling establishment: a dimmer switch‐regulated process between dark and light signaling. Plant Physiol. 176, 1061–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, L. , Wang, Z.Y. , Lin, H. , Cui, W.E. , Chen, J. , Liu, M.H. , Chen, Z.L. et al. (2006) Expression and functional analysis of the rice plasma‐membrane intrinsic protein gene family. Cell Res. 16, 277–286. [DOI] [PubMed] [Google Scholar]
- Hammerschmidt, M. , Brook, A. and McMahon, A.P. (1997) The world according to hedgehog. Trends Genet. 13, 14–21. [DOI] [PubMed] [Google Scholar]
- He, D. and Yang, P.F. (2013) Proteomics of rice seed germination. Front. Plant Sci. 4, 246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y.Q. , Yang, B. , He, Y. , Zhan, C.F. , Cheng, Y.H. , Zhang, J.H. , Zhang, H.S. et al. (2019) A quantitative trait locus, qSE3, promotes seed germination and seedling establishment under salinity stress in rice. Plant J. 97, 1089–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y.Q. , Zhao, J. , Yang, B. , Sun, S. , Peng, L.L. and Wang, Z.F. (2020) Indole‐3‐acetate beta‐glucosyltransferase OsIAGLU regulates seed vigour through mediating crosstalk between auxin and abscisic acid in rice. Plant Biotechnol. J. 18, 1933–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei, Y. , Ohta, S. , Komari, T. and Kumashiro, T. (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T‐DNA. Plant J. 6, 271–282. [DOI] [PubMed] [Google Scholar]
- Jang, J.Y. , Kim, D.G. , Kim, Y.O. , Kim, J.S. and Kang, H. (2004) An expression analysis of a gene family encoding plasma membrane aquaporins in response to abiotic stresses in Arabidopsis thaliana . Plant Mol. Biol. 54, 713–725. [DOI] [PubMed] [Google Scholar]
- Jiang, J. (2021) Hedgehog signaling mechanism and role in cancer. Sem. Cancer Biol. S1044‐579X(21)00104‐8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, N. , Shi, S. , Shi, H. , Khanzada, H. , Wassan, G.M. , Zhu, C. , Peng, X. et al. (2017) Mapping QTL for seed germinability under low temperature using a new high‐density genetic map of rice. Front. Plant Sci. 8, 1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang, W. , Lee, J. , Jin, Y.M. , Qiao, Y. , Piao, R. , Jang, S.M. , Woo, M.O. et al. (2011) Identification of QTLs for seed germination capability after various storage periods using two RIL populations in rice. Mol. Cells, 31, 385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, J.Y. , Choi, H.I. , Im, M.Y. and Kim, S.Y. (2002) Arabidopsis basic leucine zipper proteins that mediate stress responsive abscisic acid signaling. Plant Cell, 14, 343–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh, Y. and Katoh, M. (2006) Comparative genomics on HHIP family orthologs. Int. J. Mol. Med. 17, 391–395. [PubMed] [Google Scholar]
- Kline, K.G. , Barrett‐Wilt, G.A. and Sussman, M.R. (2010) In planta changes in protein phosphorylation induced by the plant hormone abscisic acid. Proc. Natl Acad. Sci. USA, 107, 15986–15991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, V. and Ladha, J.K. (2011) Direct seeding of rice: recent developments and future research needs. Adv. Agron. 111, 297–413. [Google Scholar]
- Kushiro, T. , Okamoto, M. , Nakabayashi, K. , Yamagishi, K. , Kitamura, S. , Asami, T. , Hirai, N. et al. (2004) The Arabidopsis cytochrome P450 CYP707A encodes ABA 8'‐hydroxylases: key enzymes in ABA catabolism. EMBO J. 23, 1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, H.H. , Ribaut, J.M. , Li, Z.L. and Wang, J.K. (2008) Inclusive composite interval mapping (ICIM) for digenic epistasis of quantitative traits in biparental populations. Theor. Appl. Genet. 116, 243–260. [DOI] [PubMed] [Google Scholar]
- Li, Z.H. , Zhang, J. , Liu, Y.L. , Zhao, J.H. , Fu, J.J. , Ren, X.L. , Wang, G.Y. et al. (2016) Exogenous auxin regulates multi‐metabolic network and embryo development, controlling seed secondary dormancy and germination in Nicotiana tabacum L. BMC Plant Biol. 16, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, C.W. , Fukumoto, T. , Matsumoto, T. , Gena, P. , Frascaria, D. , Kaneko, T. , Katsuhara, M. et al. (2013) Aquaporin OsPIP1;1 promotes rice salt resistance and seed germination. Plant Physiol. Biochem. 63, 151–158. [DOI] [PubMed] [Google Scholar]
- Liu, H.Y. , Hussain, S. , Zheng, M.M. , Peng, S.B. , Huang, J.L. , Cui, K.H. and Nie, L.X. (2015) Dry direct‐seeded rice as an alternative to transplanted‐flooded rice in central China. Agron Sustain. Dev. 35, 285–294. [Google Scholar]
- Liu, H.Y. , Yu, X. , Cui, D.Y. , Sun, M.H. , Sun, W.N. , Tang, Z.C. , Kwak, S.S. et al. (2007) The role of water channel proteins and nitric oxide signaling in rice seed germination. Cell Res. 17, 638–649. [DOI] [PubMed] [Google Scholar]
- Liu, L.F. , Lai, Y.Y. , Cheng, J.P. , Wang, L. , Du, W.L. , Wang, Z.F. and Zhang, H.S. (2014) Dynamic quantitative trait locus analysis of seed vigor at three maturity stages in rice. PLoS One, 9, e115732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez‐Molina, L. , Mongrand, S. and Chua, N.H. (2001) A post germination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis . Proc. Natl Acad. Sci. USA, 98, 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, X.F. , Dai, Y.J. , Zheng, C. , Yang, Y.Z. , Chen, W. , Wang, Q.C. , Chandrasekaran, U. et al. (2021) The ABI4‐RbohD/VTC2 regulatory module promotes reactive oxygen species (ROS) accumulation to decrease seed germination under salinity stress. New Phytol. 229, 950–962. [DOI] [PubMed] [Google Scholar]
- Ma, Y. , Szostkiewicz, I. , Korte, A. , Moes, D. , Yang, Y. , Christmann, A. and Grill, E. (2009) Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science, 324, 1064–1068. [DOI] [PubMed] [Google Scholar]
- Mahender, A. , Anandan, A. and Pradhan, S.K. (2015) Early seedling vigour, an imperative trait for direct‐seeded rice, an overview on physio‐morphological parameters and molecular markers. Planta, 241, 1027–1050. [DOI] [PubMed] [Google Scholar]
- Mariaux, J.B. , Bockel, C. , Salamini, F. and Bartels, D. (1998) Desiccation‐ and abscisic acid‐responsive genes encoding major intrinsic proteins (MIPs) from the resurrection plant Craterostigma plantagineum . Plant Mol. Biol. 38, 1089–1099. [DOI] [PubMed] [Google Scholar]
- McCouch, S.R. , Wright, M.H. , Tung, C.‐W. , Maron, L.G. , McNally, K.L. , Fitzgerald, M. , Singh, N. et al. (2016) Open access resources for genome‐wide association mapping in rice. Nat. Commun. 7, 10532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray, M.G. and Thompson, W.F. (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara, E. , Keith, K. , McCourt, P. and Naito, S. (1995) A regulatory role for the ABI3 gene in the establishment of embryo maturation in Arabidopsis thaliana . Development, 121, 629–636. [Google Scholar]
- Ni, B.R. and Bradford, K.J. (1992) Quantitative models characterizing seed germination responses to abscisic Acid and osmoticum. Plant Physiol. 98, 1057–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonogaki, H. (2019) ABA responses during seed development and germination. Adv. Bot Res. 92, 171–217. [Google Scholar]
- Palaniyandi, S.A. , Chung, G. , Kim, S.H. and Yang, S.H. (2015) Molecular cloning and characterization of the ABA‐specific glucosyltransferase gene from bean (Phaseolus vulgaris L.). J. Plant Physiol. 178, 1–9. [DOI] [PubMed] [Google Scholar]
- Park, S.‐Y. , Fung, P. , Nishimura, N. , Jensen, D.R. , Fujii, H. , Zhao, Y. , Lumba, S. et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science, 324, 1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield, S. (2017) Seed dormancy and germination. Curr. Biol. 27, R874–R878. [DOI] [PubMed] [Google Scholar]
- Rajjou, L. , Duval, M. , Gallardo, K. , Catusse, J. , Bally, J. , Job, C. and Job, D. (2012) Seed germination and vigor. Annu. Rev. Plant Biol. 63, 507–533. [DOI] [PubMed] [Google Scholar]
- Rallis, A. , Navarro, J.A. , Rass, M. , Hu, A. , Birman, S. , Schneuwly, S. and Thérond, P.P. (2020) Hedgehog signaling modulates glial proteostasis and lifespan. Cell Rep. 30, 2627–2643.e5. [DOI] [PubMed] [Google Scholar]
- Rao, A.N. , Johnson, D.E. , Sivaprasad, B. , Ladha, J.K. and Mortimer, A.M. (2007) Weed management in direct‐seeded rice. Adv. Agron. 93, 153–255. [Google Scholar]
- Roche, J.V. and Törnroth‐Horsefield, S. (2017) Aquaporin protein‐protein interactions. Int. J. Mol. Sci. 18, 2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano, N. and Marion‐Poll, A. (2021) ABA metabolism and homeostasis in seed dormancy and germination. Int. J. Mol. Sci. 22, 5069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seiler, C. , Harshavardhan, V.T. , Rajesh, K. , Reddy, P.S. , Strickert, M. , Rolletschek, H. , Scholz, U. et al. (2011) ABA biosynthesis and degradation contributing to ABA homeostasis during barley seed development under control and terminal drought‐stress conditions. J. Exp. Bot. 62, 2615–2632. [DOI] [PubMed] [Google Scholar]
- Sherrier, D.J. , Prime, T.A. and Dupree, P. (1999) Glycosylphosphatidylinositol‐anchored cell‐surface proteins from Arabidopsis . Electrophoresis, 20, 2027–2035. [DOI] [PubMed] [Google Scholar]
- Shu, K. , Liu, X.D. , Xie, Q. and He, Z.H. (2016) Two faces of one seed: hormonal regulation of dormancy and germination. Mol. Plant, 9, 34–45. [DOI] [PubMed] [Google Scholar]
- Shu, K. , Zhang, H.W. , Wang, S.F. , Chen, M.L. , Wu, Y.R. , Tang, S.Y. , Liu, C.Y. et al. (2013) ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis . PLoS Genet. 9, e1003577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderman, E.M. , Brocard, I.M. , Lynch, T.J. and Finkelstein, R.R. (2000) Regulation and function of the Arabidopsis ABA‐insensitive 4 gene in seed and abscisic acid response signaling networks. Plant Physiol. 124, 1752–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J.W. , Shang, L.L. , Wang, X. , Xing, Y.L. , Xu, W. , Zhang, Y.Y. , Wang, T.T. et al. (2021) MAPK11 regulates seed germination and ABA signaling in tomato by phosphorylating SnRKs. J. Exp. Bot. 72, 1677–1690. [DOI] [PubMed] [Google Scholar]
- Suga, S. , Komatsu, S. and Maeshima, M. (2002) Aquaporin isoforms responsive to salt and water stresses and phytohormones in radish seedlings. Plant Cell Physiol. 43, 1229–1237. [DOI] [PubMed] [Google Scholar]
- Tabin, C.J. and McMahon, A.P. (1997) Recent advances in Hedgehog signaling. Trends Cell Biol. 7, 442–446. [DOI] [PubMed] [Google Scholar]
- Vander Willigen, C. , Postaire, O. , Tournaire‐Roux, C. , Boursiac, Y. and Maurel, C. (2006) Expression and inhibition of aquaporins in germinating Arabidopsis seeds. Plant Cell Physiol. 47, 1241–1250. [DOI] [PubMed] [Google Scholar]
- Wang, Z.F. , Wang, J.F. , Bao, Y.M. , Wang, F.H. and Zhang, H.S. (2010) Quantitative trait loci analysis for rice seed vigor during the germination stage. J. Zhejiang Univ. Sci. B, 11, 958–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, L.X. , Tan, Z.W. , Zhou, Y. , Xu, R.B. , Feng, L.B. , Xing, Y.Z. and Qi, X.Q. (2014) Identification and fine mapping of quantitative trait loci for seed vigor in germination and seedling establishment in rice. J. Integr. Plant Biol. 56, 749–759. [DOI] [PubMed] [Google Scholar]
- Xing, H.L. , Dong, L. , Wang, Z.P. , Zhang, H.Y. , Han, C.Y. , Liu, B. , Wang, X.C. et al. (2014) A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 14, 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, E.S. , Chen, M.M. , He, H.Y. , Zhan, C.F. , Cheng, Y.H. , Zhang, H.S. and Wang, Z.F. (2017) Proteomic Analysis reveals proteins involved in seed imbibition under salt stress in rice. Front. Plant Sci. 7, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelazny, E. , Borst, J.W. , Muylaert, M. , Batoko, H. , Hemminga, M.A. and Chaumont, F. (2007) FRET imaging in living maize cells reveals that plasma membrane aquaporins interact to regulate their subcellular localization. Proc. Natl Acad. Sci. USA, 104, 12359–12364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, P. , Zhu, P.W. , Qian, L.F. , Qian, X.M. , Mi, Y.X. , Lin, Z.F. , Dong, S.N. et al. (2021) Identification and fine mapping of qGR6.2, a novel locus controlling rice seed germination under salt stress. BMC Plant Biol. 21, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X. , Garreton, V. and Chua, N.H. (2005) The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 19, 1532–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, C. , Schraut, D. , Hartung, W. and Schäffner, A.R. (2005) Differential responses of maize MIP genes to salt stress and ABA. J. Exp. Bot. 56, 2971–2981. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Comparison of seed germination and seedling establishment between JCQ and IR26 under normal condition.
Figure S2 QTLs analysis of seed germination traits among F2 population under normal condition.
Figure S3 Comparison of coding sequences in ORF2 between IR26 and Jiucaiqing.
Figure S4 Comparison of seed vigour between NIL‐qsv3 and IR26 under normal condition.
Figure S5 Comparisons of the OsHIPL1 transcript levels between transgenic lines (OE1, OE2, COM‐1 and COM‐2) and wild type (NIL‐qsv3 and ZH11).
Figure S6 The confirmation of differential expression genes (DEGs) between IR26 and NIL‐qsv3 in 12h imbibed seeds via the RT‐qPCR approach in rice.
Figure S7 GO enrichment analysis for differentially expressed genes (DEGs) in IR26 compared to NIL‐qsv3.
Figure S8 Rice OsHIPL1 altering expressions of genes involved in ABA signalling during seed germination.
Figure S9 Transcription levels of OsHIPL1 response to ABA treatments in germinating seeds conducted using the RT‐qPCR approach.
Figure S10 Comparison of seedling establishment and seedling growth between wild‐type and the transgenic lines when seeds are directly sown in outdoor field soil.
Figure S11 Comparison of agronomic traits among IR26, Oshipl1 mutants, NIL‐qsv3 and COM lines.
Figure S12 Comparison of seed germination percentage and seedling percentage between NIL‐qsv3 and IR26 under low temperature (A–C), salinity (D–F), PEG (G–I) and mannitol (J–L) stresses.
Figure S13 Comparison of seed germination percentage and seedling percentage of the transgenic lines of OsHIPL1 under salinity (A‐D) and PEG (E‐H) stresses.
Figure S14 Phylogenetic relationships of OsHIPL1 among Oryza sativa, Maize, Sorghum, Setaria italica, Setaria viridis, Brachypodium, Brachypodium stacei, Arabidopsis, Prunus persica and Glycine max.
Figure S15 The contents of IAA in germinated seeds of Oshipl1 mutants and IR26.
Table S1 The information of phenotype of seed vigour of the CSSLs and parents.
Table S2 The information of genotype of seed vigour of the CSSLs and parents.
Table S3 The information QTLs of seed vigour identified by single marker mapping based on the CSSL population.
Table S4 Differential expression genes (DEGs) between IR26 and NIL‐qsv3 in 12h imbibed seeds in rice.
Table S5 DEGs involved in hormone metabolism between IR26 and NIL‐qsv3 in 12h imbibed seeds in rice.
Table S6 Identification of putative OsHIPL1‐interacting proteins using yeast two‐hybrid screening.
Table S7 Information of the 192 accessions used for haplotype analyses in rice.
Table S8 The primer pairs used in this study.


