Figure 6.
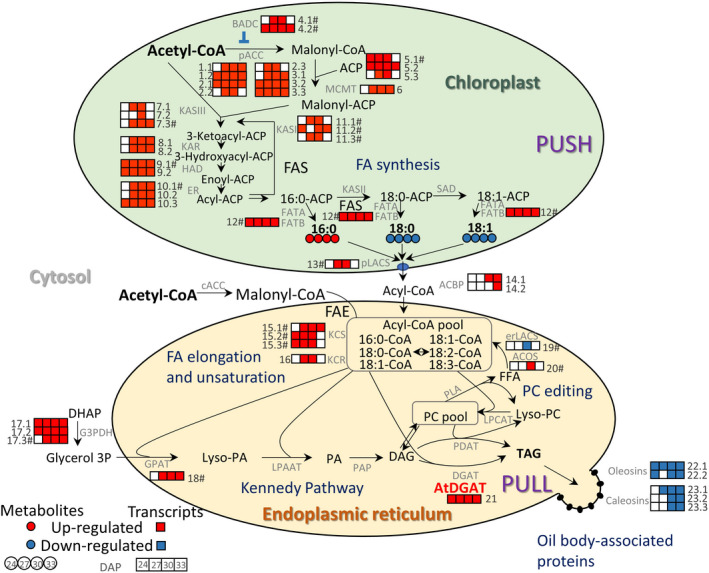
Alteration of lipid biosynthesis in WD embryos between 24 and 33 DAP. Red and blue circles were used to account for significantly increased or decreased levels of each FA at each stage, respectively (n = 4, P < 0.05). Red and blue boxes were used to show the significantly up‐ or down‐regulated expression of each enzyme at each stage under greenhouse conditions, respectively (n = 3, P < 0.05, |log2(Fold Change)| ≥ 1). The symbol # next to the corresponding enzyme number denotes the presence of the AW‐box motif in the promoter of the enzyme’s gene. ACP, plastidic acyl carrier protein; FAS, fatty acid synthase; pACC, heteromeric acetyl CoA carboxylase, biotin carboxylase subunit: (1.1) Glyma.05G221100 and (1.2) Glyma.08G027600, biotin carboxyl carrier protein: (2.1) Glyma.13G057400, (2.2) Glyma.18G265300 and (2.3) Glyma.19G028800, carboxyltransferase alpha subunit: (3.1) Glyma.18G195700, (3.2) Glyma.18G195900 and (3.3) Glyma.18G196000; BADC, biotin/lipoyl attachment domain‐containing protein: (4.1) Glyma.11G233700# and (4.2) Glyma.18G023300#; ACP, acyl carrier protein: (5.1) Glyma.13G214600#, (5.2) Glyma.15G098500 and (5.3) Glyma.20G230100; MCMT, malonyl‐CoA: ACP malonyltransferase: (6) Glyma.11G164500; KASIII, ketoacyl‐ACP synthase III: (7.1) Glyma.09G277400, (7.2) Glyma.15G003100 and (7.3) Glyma.18G211400#; KAR, ketoacyl‐ACP reductase: (8.1) Glyma.11G248000 and (8.2) Glyma.18G009200; HAD, hydroxyacyl‐ACP dehydrase: (9.1) Glyma.08G179900# and (9.2) Glyma.15G052500; ER, enoyl‐ACP reductase: (10.1) Glyma.08G345900#, (10.2) Glyma.12G027300, and (10.3) Glyma.18G156100; KASI, ketoacyl‐ACP synthase I: (11.1) Glyma.05G129600#, (11.2) Glyma.08G024700# and (11.3) Glyma.08G084300#; SAD, stearoyl‐ACP desaturase; FATA, fatty acyl thioesterase A; FATB, fatty acyl thioesterase B: (12) Glyma.04G151600#; pLACS, plastidic long‐chain acyl‐ CoA synthetase: (13) Glyma.06G112900#; ACBP, acyl CoA‐binding protein: (14.1) Glyma.13G152900 and (14.2) Glyma.11G014900; ACCc, homomeric acetyl CoA carboxylase; FAE, fatty acid elongase; KCS, ketoacyl‐CoA synthase: (15.1) Glyma.06G012500#, (15.2) Glyma.04G149300# and (15.3) Glyma.06G214800#; KCR, ketoacyl‐CoA reductase: (16) Glyma.11G245600; DHAP, dihydroxyacetone phosphate; G3PDH, glycerol 3 phosphate dehydrogenase: (17.1) Glyma.19G053500, (17.2) Glyma.02G186600 and (17.3) Glyma.10G107100#; GPAT, glycerol‐3‐phosphate acyltransferase: (18) Glyma.20G070400; Lyso‐PA, lysophosphatidic acid; LPAAT, 2‐lysophosphatidic acid acyltransferase; PA, phosphatidic acid; PAP, phosphatidate phosphatase; DAG, diacylglycerol; TAG, triacylglycerol; DGAT, acyl‐CoA: diacylglycerol acyltransferase; PDAT, phospholipid:diacylglycerol acyltransferase; PC, phosphatidylcholine; LPCAT, lysophosphatidylcholine acyltransferase; PLA, phospholipase A; erLACS, ER long‐chain acyl‐ CoA synthetase: (19) Glyma.10G010800#; ACOS, acyl‐CoA synthase: (20) Glyma.08G329700; FFA, free fatty acid; AtDGAT, transgen expressed (21); Oleosin: (22.1) Glyma.05G004300 and (22.2) Glyma.19G004800; and Caleosins: (23.1) Glyma.10G189900, (23.2) Glyma.20G200900, (23.3) Glyma.20G201000. DAP: days after pollination.
