Figure 7.
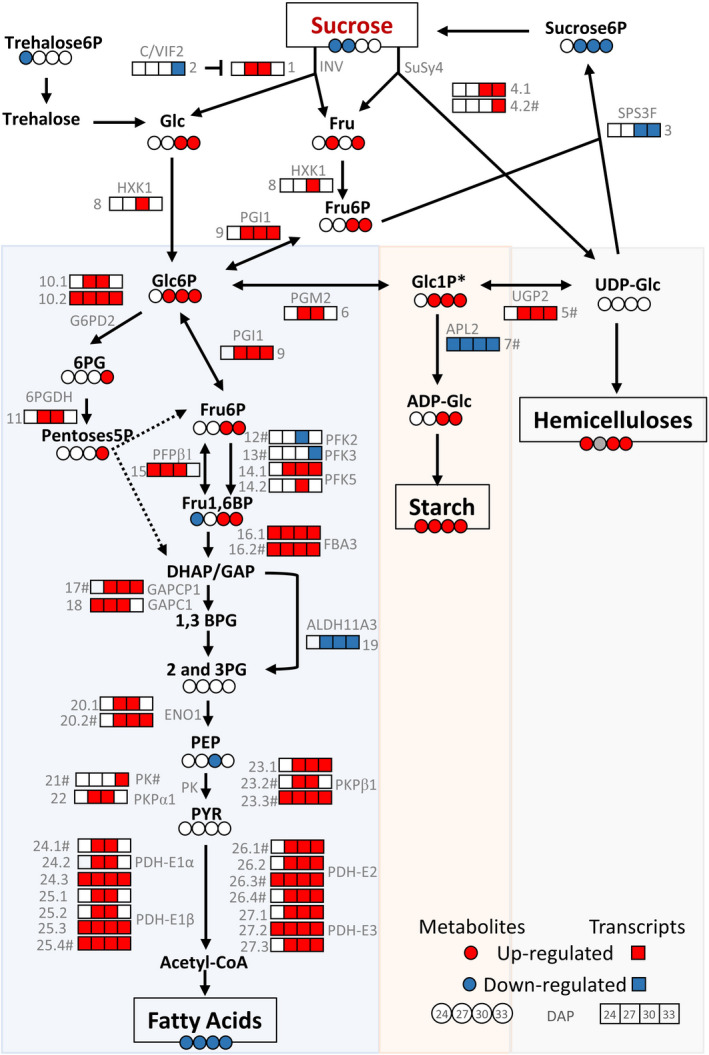
Carbon partitioning in WD embryos between 24 and 33 DAP. Metabolites and end products: red and blue circles were used to account for significantly increased or decreased levels of each metabolite at each stage, respectively (n = 4, P < 0.05) and a grey circle reflects data not measured. The contents of Glc1P are shown with an asterisk (*) as were determined together with the levels of mannose 1‐phosphate. Sucrose6P, sucrose 6‐phosphate; Threhalose6P, trehalose 6‐phosphate; Glc, glucose; Fru, fructose; Glc6P, glucose 6‐phosphate; Glc1P, glucose 1‐phosphate; Fru6P, fructose 6‐phosphate; Fru1,6BP, fructose 1,6‐bisphosphate; UDP‐Glc, UDP‐glucose; ADP‐Glc, ADP‐glucose; 6PG, 6‐phosphogluconate; Pentoses5P, pentoses 5‐phosphate; DHAP, dihydroxyacetone phosphate; GAP, glyceraldehyde 3 phosphate; 1,3BPG, 1,3‐bisphosphoglycerate; 2 and 3PG, 2‐and 3‐phosphoglycerate; PEP, phosphoenolpyruvate and PYR, pyruvate. Enzyme expression: red and blue boxes were used to show the significantly up‐ or down‐regulated expression of each enzyme at each stage under greenhouse conditions, respectively (n = 3, P < 0.05). The enzyme names correspond to the specific homolog isoforms in Arabidopsis. The symbol # denotes the presence of the AW‐box motive in the promoter of the enzyme’s gene. INV, neutral invertase (1) Glyma.04G005700; C/VIF2, cell wall/vacuolar inhibitor of fructosidase 2 (2) Glyma.04G214000; SPS3F, sucrose phosphate synthase 3F (3) Glyma.18G108100; SuSy4, sucrose synthase 4 (4.1) Glyma.09G073600 and (4.2) Glyma.13G114000#; UGP2, UDP‐glucose pyrophosphorylase 2 (5) Glyma.13G152500#; PGM2, phosphoglucomutase (6) Glyma.08G044100; APL2, ADP‐glucose pyrophosphorylase large subunit (7) Glyma.14G072500#; HXK1, hexokinase 1 (8) Glyma.07G015100; PGI1, phosphoglucose isomerase 1 (9) Glyma.06G094300;G6PD2, glucose 6 phosphate dehydrogenase 2 (10.1) Glyma.02G096800 and (10.2) Glyma.18G284600; 6PGDH, 6‐phosphogluconate dehydrogenase (11) Glyma.05G214000; PFK2, phosphofructokinase 2 (12) Glyma.07G269500#;PFK3, phosphofructokinase 3 (13) Glyma.13G353400#; PFK5, phosphofructokinase 5 (14.1) Glyma.08G280700 and (14.2) Glyma.18G145500; PFPβ1, pyrophosphate: fructose 6‐phosphate 1‐phosphotransferase (15) Glyma.18G135100; FBA3, fructose bisphosphate aldolase 3 (16.1) Glyma.10G268500 and (16.2) Glyma.20G122500#; GAPCP1, glyceraldehyde‐3‐phosphate dehydrogenase of plastid 1 (17) Glyma.03G092700#; GAPC1, glyceraldehyde 3 phosphate dehydrogenase C subunit 1 (18) Glyma.11G247600; ALDH11A3, NADP‐dependent glyceraldehyde 3‐phosphate dehydrogenase (19) Glyma.02G202500; ENO1, enolase 1 (20.1) Glyma.04G144300 and (20.2) Glyma.06G208200#; PK, pyruvate kinase family protein (21) Glyma.19G190100;PKPα1, plastidial pyruvate kinase 1 subunit alpha (22) Glyma.20G211200; PKPβ1, plastidial pyruvate kinase 1 subunit beta (23.1) Glyma.09G126300, (23.2) Glyma.10G227800#, (23.3) Glyma.16G173100#; PDH‐E1α, E1a component of pyruvate dehydrogenase complex (24.1) Glyma.03G261000#, (24.2) Glyma.16G018300, (24.3) Glyma.19G260000; PDH‐E1β, E1b component of pyruvate dehydrogenase complex (25.1) Glyma.05G141000, (25.2) Glyma.08G096300, (25.3) Glyma.14G092100, (25.4) Glyma.17G231400#; PDH‐E2, E2 component of pyruvate dehydrogenase complex (26.1) Glyma.10G215400#, (26.2) Glyma.20G176300, (26.3) Glyma.01G081900#, (26.4) Glyma.20G114500# and PDH‐E3, E3 component of pyruvate dehydrogenase complex (27.1) Glyma.15G143100, (27.2) Glyma.07G241600, (27.3) Glyma.17G032300.
