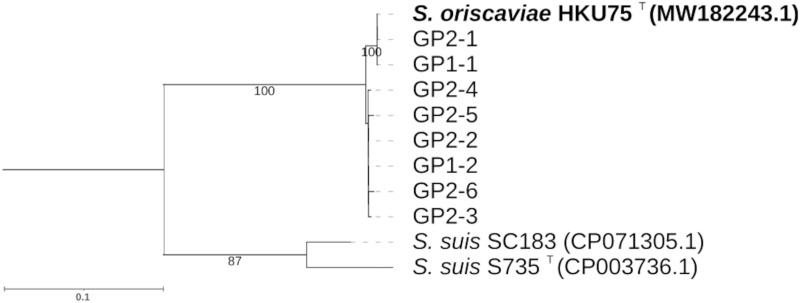
FIG 2.
Phylogenetic analysis of groEL sequences detected in the oral swabs of guinea pigs GP-1 and GP-2. The tree was constructed by the maximum-likelihood method using the Tamura 3-parameter model with L. lactis ATCC 19435T as the outgroup. A total of 588 nucleotide positions was included in the analysis. Bootstrap values were calculated as percentages from 1,000 pseudoreplicates (values lower than 70 are not shown). The scale bar indicates the estimated number of substitutions per 100 bases. Names and GenBank nucleotide accession numbers are given as cited in GenBank. The accession number for the groEL gene sequence of L. lactis ATCC 19435T is FMTF01000003.