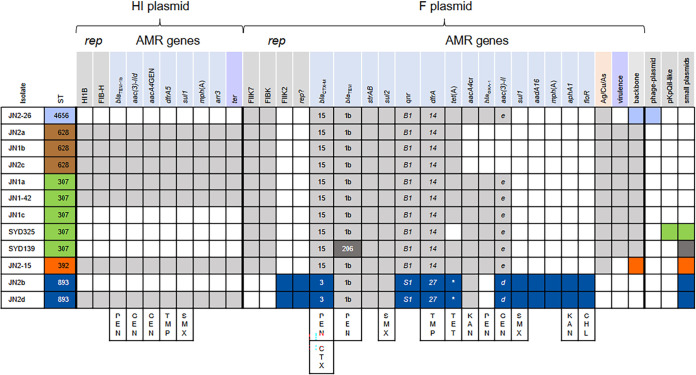
FIG 4.
Correlation of antibiotic resistance gene content, plasmid replicons and predicted resistance phenotypes. rep, all from PlasmidFinder (59); HIB 567/570 nucleotides identical to “IncHI1B(pNDM-MAR)”; FIB-H, identical to “IncFIB(pNDM-MAR)”; FIIK7, FIIK-type replicon, K7 by RST (59); FIB(K), FIB(K)-type replicon; FIIK2, FII(K)-type replicon, K2 by RST; rep?, replication initiation gene of unnamed type. Antibiotic resistance (AMR) genes are organized according to proposed plasmid location (as shown in Fig. S2 and S3). Gene variants identified by mapping raw reads are specified. *, in the tet(A) column indicates that the last 9 amino acids of the Tet(A) protein are different due to truncation of the tet(A) gene. ter, region associated with resistance to stress induced by resident gut microflora (28); Ag/Cu/As, silver, copper and arsenic resistance regions; virulence, urea ABC-transport, Fec-like iron (III) dicitrate transport, glutathione ABC-transporter; lacIZY, glycogen synthesis; backbone, backbone type (see Fig. S2); phage-plasmid, phage element with a region 95% identical to PlasmdFinder “IncFIB(pKPHS1)” target; pKpQIL, pKpQIL-like plasmid carrying blaKPC-3. Last column, small plasmids were found as circular contigs: SYD325, 34.032 kb N3-type plasmid plus 1.546 kb plasmid; SYD139, 3.991 kb plasmid (~40 copies/chromosome) matching one found in K. pneumoniae, and 3.223 kb plasmid (~37 copies/chromosome), closely related to plasmids found in various Enterobacterales; JN2-15, 1.552 kb plasmid (~50 copies/chromosome) identical to plasmids found mainly in E. coli; JN2b and JN2d, 5.617 kb plasmid with no close matches and 5.251 kb plasmid almost identical to plasmids from K. pneumoniae and other species (both ~60 copies/chromosome). Relevant resistance phenotypes conferred by selected resistance genes are shown at the bottom: PEN, penicillin; GEN, gentamicin; TMP, trimethoprim; SMX, sulfamethoxazole; CTX, cefotaxime; TET, tetracycline; KAN, kanamycin; CHL, chloramphenicol. blaCTX-M-15 would be expected to confer resistance to ceftazidime in addition to CTX, while blaCTX-M-3 would not. qnrB1, qnrS1 and aacA4cr could all contribute to fluoroquinolone resistance.