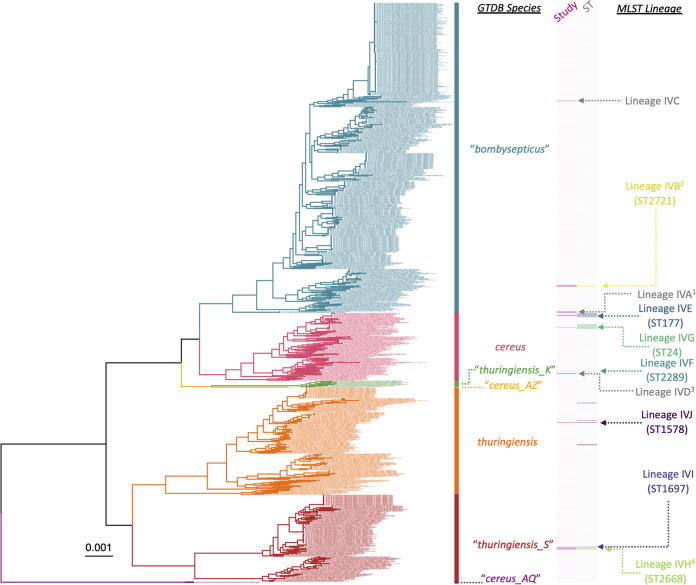
FIG 3.
Maximum likelihood (ML) phylogeny constructed using core SNPs identified among orthologous gene clusters of 1,081 panC group IV B. cereus sensu lato genomes. The phylogeny was rooted using panC group III B. anthracis strain Ames Ancestor as an outgroup (NCBI RefSeq accession number GCF_000008445.1; omitted for readability). Tip label colors and clade labels denote species assigned using GTDB-Tk (“GTDB Species”). The heat map to the right of the phylogeny denotes (i) whether a strain was sequenced in this study (dark pink) or not (light pink; “Study”) and (ii) multilocus sequence typing (MLST) sequence types (STs) associated with strains sequenced in this study, where applicable (colored), or not (gray; “ST”). MLST lineages discussed in Table 3 are annotated to the right of the heat map (“MLST Lineage”). MLST lineages with numerical superscripts contain two or more strains sequenced in this study, which were highly similar on a genomic scale; these lineages are depicted in Fig. 4. Branch lengths are reported in substitutions per site.