FIG 1.
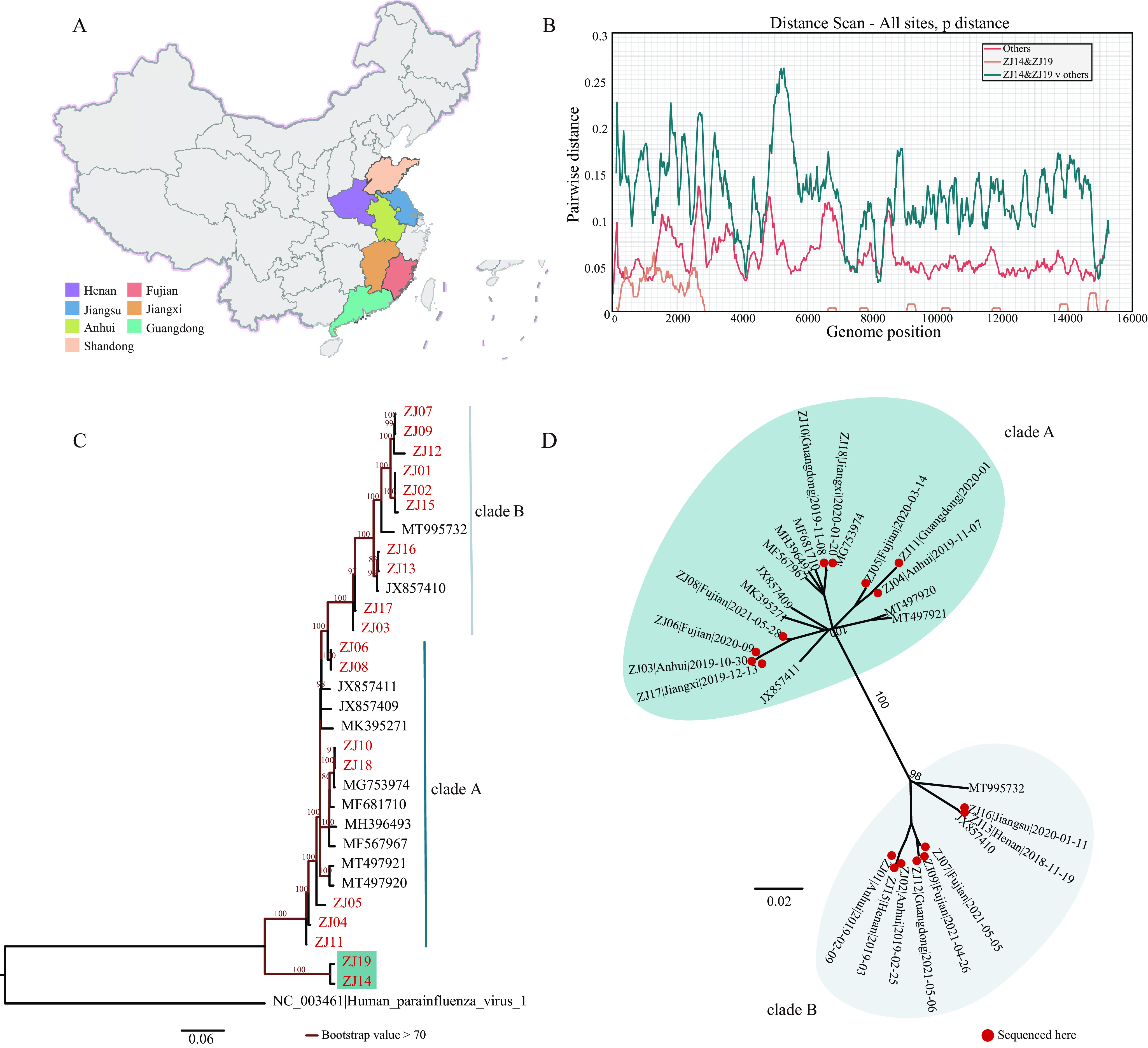
Diversity of newly sequenced porcine paramyxovirus. (A) Provinces of the PPIV strains sequenced in this study. (B) Pairwise distance of the whole genomes of newly sequenced porcine paramyxovirus and available PPIV-1 sequences. We used SSE1.4 with a sliding window of 250 nucleotides, and a step size of 25 nucleotides to plot the pairwise distance. Different colored lines represent different groups. (C) The reconstructed maximum-likelihood phylogenetic tree based on the nucleotide sequences of L gene with HPIV-1 as the outer group. The unique branches formed by ZJ14 and ZJ19 are shown in blue green. (D) The reconstructed maximum-likelihood phylogeny of PPIV-1 whole genome, after removing ZJ14 and ZJ19. Strains in colored regions represent the different clades of PPIV-1. The red dots represent the strains sequenced in this study.
