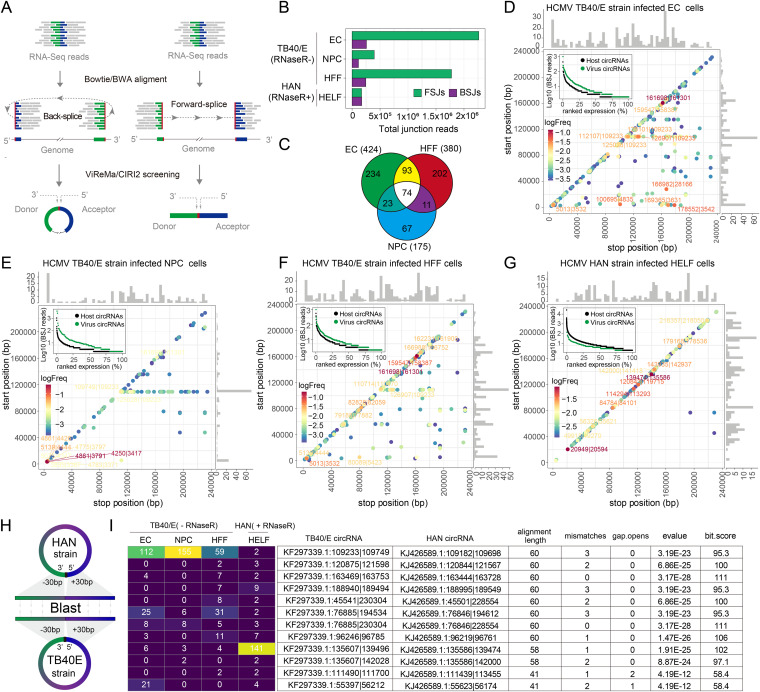
FIG 1.
Predicted HCMV circRNAs and their abundances and landscapes. (A) Schematic diagram of CIRI2-based identification of circRNAs. Junction reads are aligned to the donor and acceptor sequences. If the genomic location of the donor is downstream of the acceptor, it is considered a back-splice junction (BSJ). The 5′ and 3′ breakpoints are then determined. (B) ViReMa-based identification of HCMV FSJs and BSJs from HCMV-infected cells. (C) Venn diagram presenting the number of unique or shared circRNAs in each paired group. (D to G) Frequency of circularization events in HCMV TB40/E-strain-infected EC (D), NPCs (E), HFF (F), and HAN-strain-infected-HELF cells (G). Counts of BSJ-spanning reads of circRNAs (starting from a coordinate on the x-axis and ending in a coordinate on the y-axis) are indicated by the colors. The numbers of circRNA start/stop positions are shown as histograms on the x-axis and y-axis. The ranked expression level of de novo identified circRNAs from HCMV and human genomes. The labels of the top 10 circRNAs are given according to their genome positions. The complete list of splice sites is provided in Data Set S2. (H) Schematic diagram of the alignment between the HAN and TB40/E strain circRNAs. (I) The alignment results of the top 12 conserved HCMV circRNAs from the infected cells.