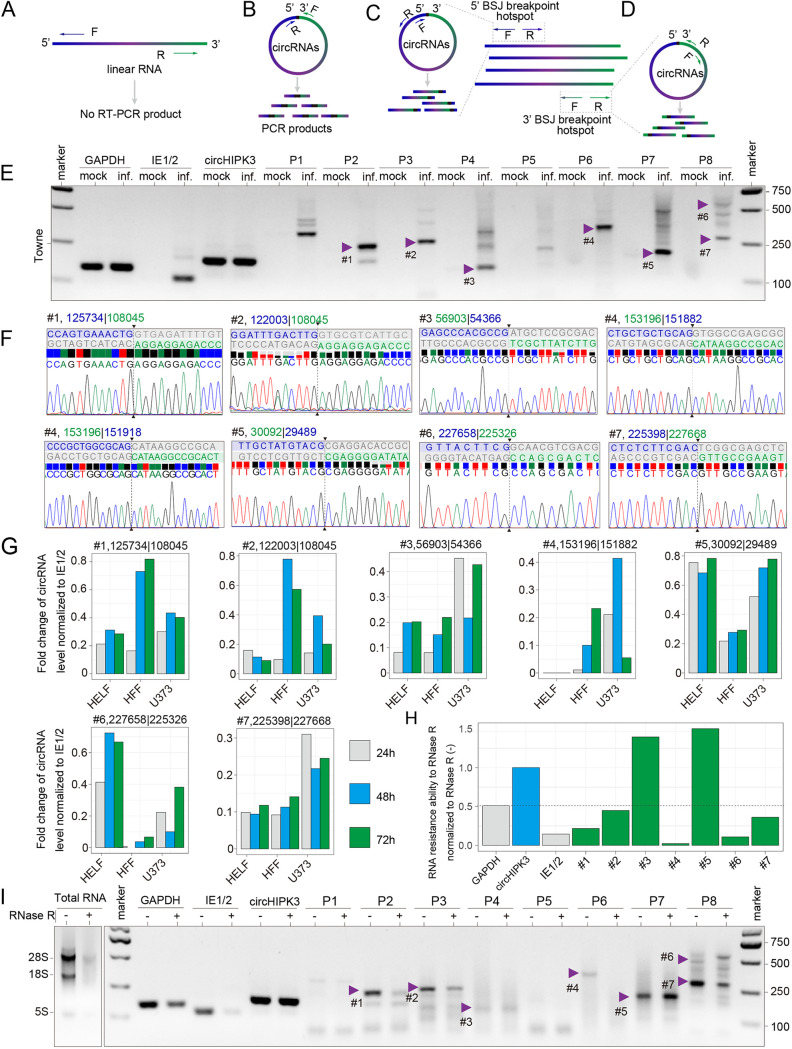
FIG 4.
Experimental identification of HCMV circRNAs. (A) Illustration of how inverse RT-PCR with divergent primers selectively amplifies different regions of circRNAs but not linear RNAs. (B to D) Schematic diagram of how divergent primers were designed to amplify predicted BSJs (B) and confirmed full-length circRNA sequences (C and D). (E) Inverse RT-PCR result of the HCMV laboratory strain Towne with primer sets shown in (B). Bands indicated by arrows were sequenced. Representative Sanger sequencing results are shown in (F). BSJ breakpoints are indicated by dashed lines. Donor (green), acceptor (blue) sequences, and downstream/upstream sequences (gray) flanking the junction were aligned with the BSJ sequence. (H and I) Relative resistance ability to RNase R of linear RNA and circRNAs (G). The relative expression of HCMV circRNAs in HELF, HFF and U373 cells at different times of infection. The values were normalized to IE1/2. The value was normalized to the corresponding sample without RNase R treatment. (H). Bands are shown in (I), where the left panel shows an agarose gel analysis of total RNA with and without RNase R treatment, and the right panel shows the products of RT-PCR on RNA treated with and without RNase R. Human linear RNA, GAPDH, and HCMV linear RNA, IE1/2 (immediate early 1/2 gene, UL123) (35 cycles with convergent primers), Human circRNA, circHIPK3, and HCMV circRNAs (35 cycles with divergent primers).