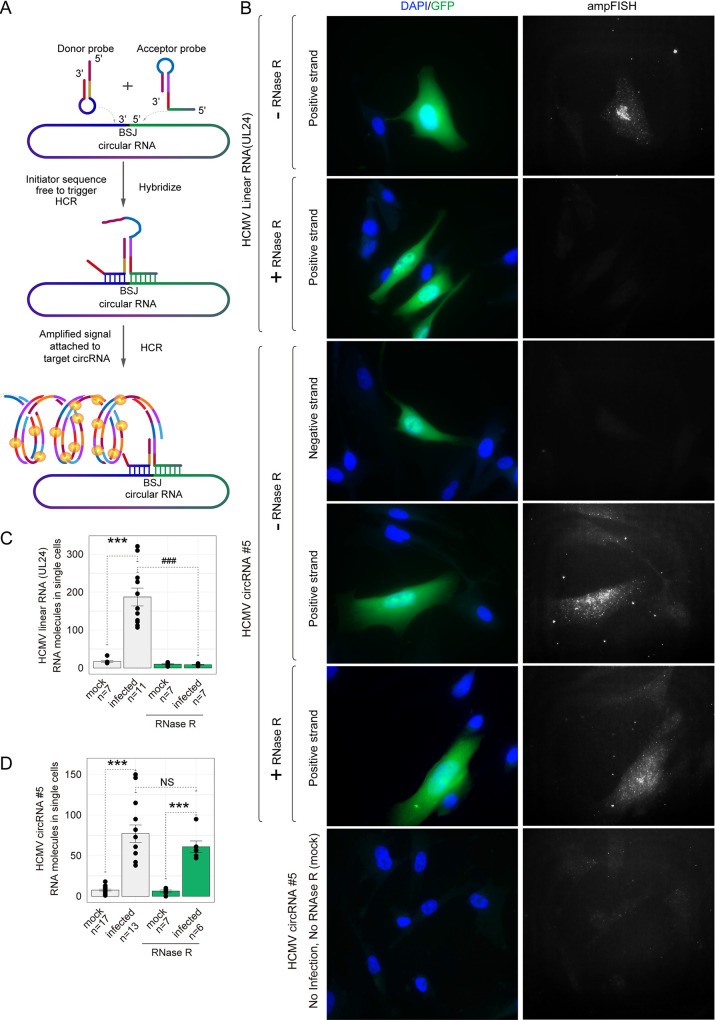
FIG 5.
Detection of an HCMV circRNA in situ using ampFISH. (A) A schematic illustrating ampFISH. When two hairpin-shaped probes bind at adjacent locations on a target RNA, conformational reorganization occurs in one of them, which initiates a hybridization chain reaction (HCR) that deposits fluorescence at the target RNA molecule. The target sequences of the two probes are present next to each other in the circular RNA but not in the linear RNAs. (B) Representative HCMV-infected MRC5 cells were imaged after ampFISH either against linear RNA UL24 or circular RNA number 5 (FJ616285.1:30092|29489). Cells harboring HCMV are GFP-positive. ampFISH images are maximal intensity merges of z-stacks acquired in the Cy5 channel and presented under a common contrast level. (C and D) The number of ampFISH spots corresponding to single mRNA molecules counted in single cells for the indicated categories of cells. The evidence of the accuracy of spot counting is presented in Fig. S4. Two-tailed unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.001 mock versus infected group; ###, P < 0.001 without RNase R treatment versus with RNase R treatment group. Data are presented as mean ± S.E.M. The number of cells analyzed ranged from 6 to 16.