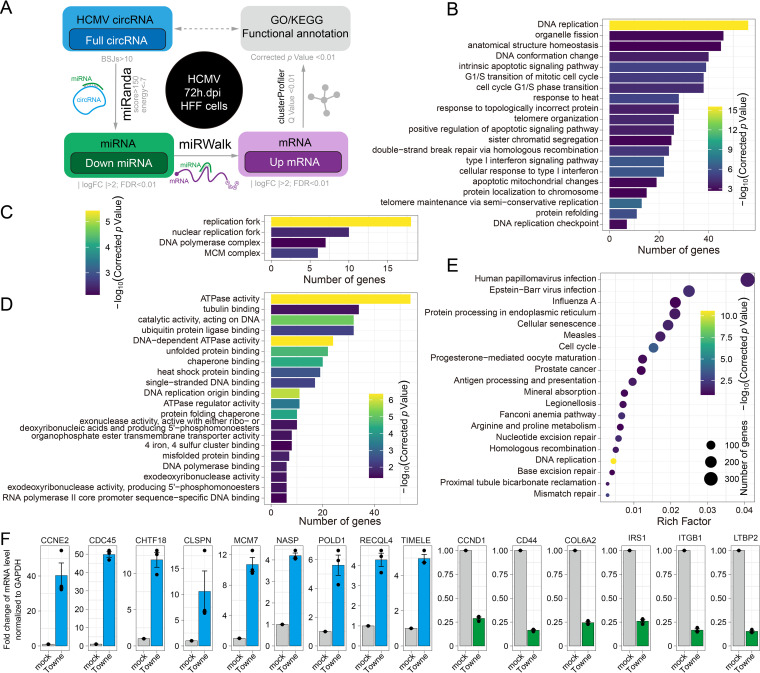
FIG 8.
Potential HCMV circRNAs function in the competitive endogenous RNA coregulatory network. (A) Schematic of potential HCMV circRNA competitive endogenous RNA coregulatory network analysis method. (B to D) Comparison of Gene Ontology (GO) enrichment of HCMV-circRNA-associated genes that are upregulated upon HCMV infection. The figure shows the top 20 significantly enriched GO terms, including biological processes (B), cellular components (C), and molecular functions (D). (E) KEGG pathways of HCMV-circRNA-associated genes that are upregulated upon HCMV infection. (F) The mRNA expression levels of HCMV-circRNA-associated genes that are upregulated upon HCMV infection (NASP, POLD1, CLSPN, MCM7, RECQL4, CHTF8, CDC45, TIMELE, and CCNE2). Genes that are downregulated upon HCMV infection (CCND1, CD44, COL6A2, IRS1, ITGB1, and LTB2) were used as negative controls using quantitative real-time PCR.