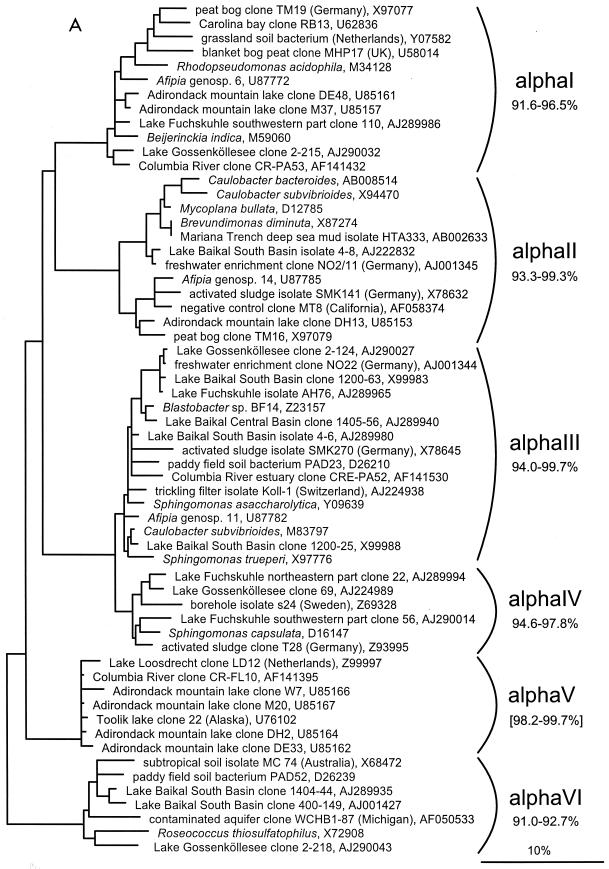
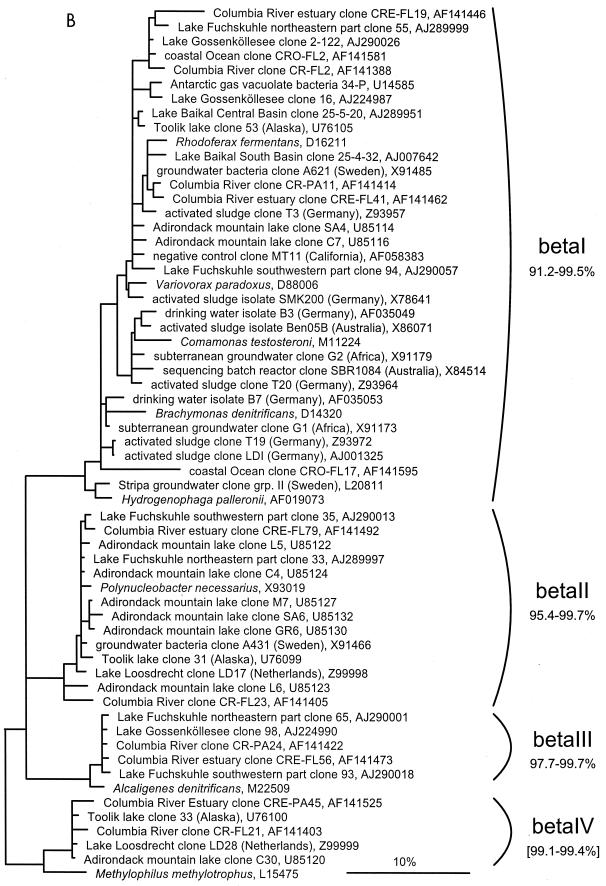
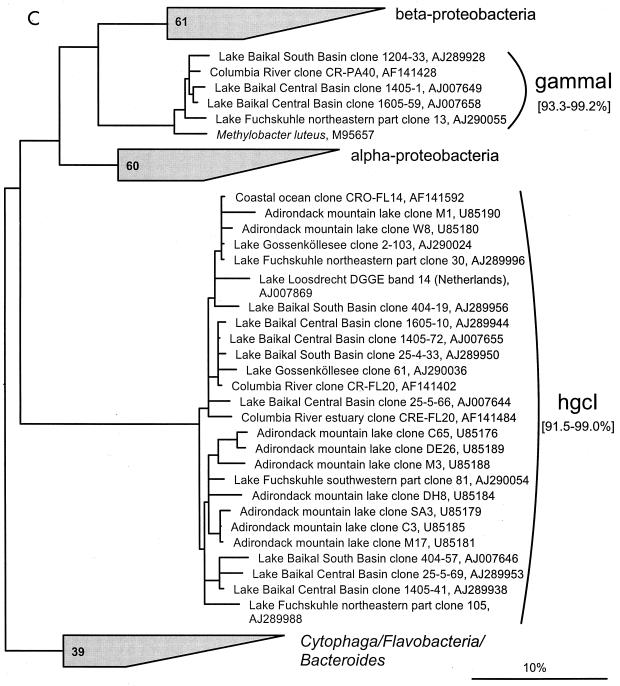
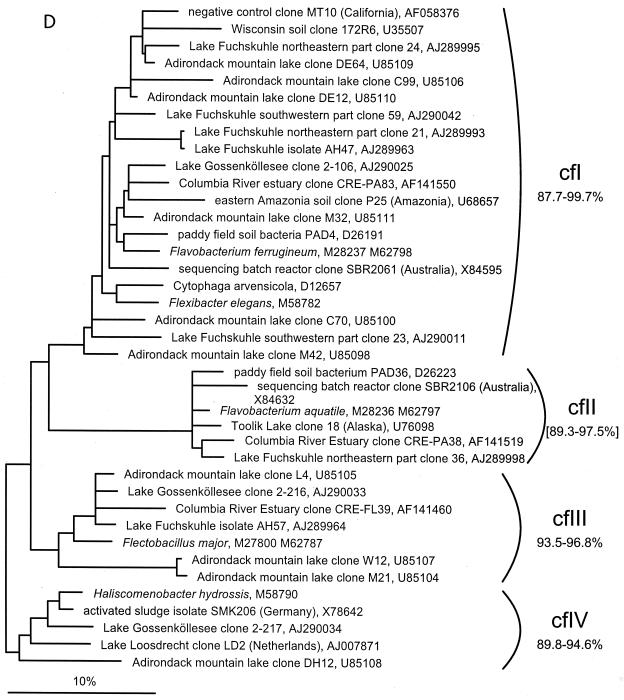
FIG. 1.
(A to D) Unrooted phylogenetic consensus tree based on maximum likelihood (FastDNAml) analysis showing the clones in the proposed clusters. (A) α-Proteobacteria; (B) β-Proteobacteria; (C) γ-Proteobacteria and Actinobacteria; (D) CFB phylum. Bifurcations indicate branchings which appeared stable and well separated from neighboring branchings in all cases. Multifurcations indicate tree topologies which could not be unambiguously resolved based on the available data set. For clarity, only the reported clusters are shown; all sequences between the clusters have been omitted. These are included in a more detailed version of the tree which is available at ftp.mpi-bremen.de/pub/molecol_p/fog-aem. The scale bar indicates 10% estimated sequence divergence.