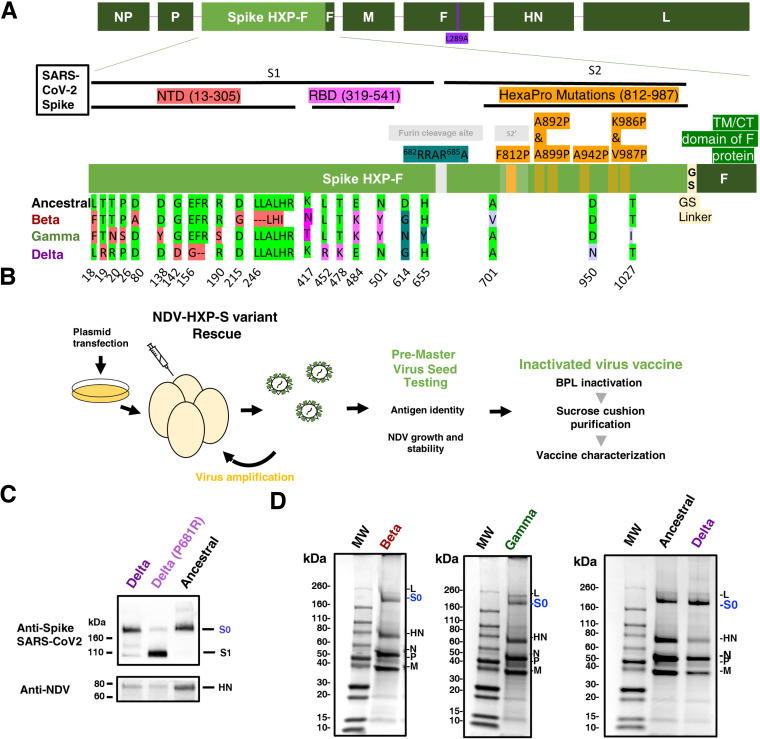
FIG 2.
Design, production, and characterization of NDV-HXP-S variant vaccines. (A) Structure and design of the NDV-HXP-S construct. The different SARS-CoV-2 spike sequences were introduced between the P and M genes of LaSota L289A NDV strain. The ectodomain of the spike was connected to the transmembrane domain and cytoplasmic tail (TM/CT) of the F protein of the NDV. The original polybasic cleavage site was removed by mutating RRAR to A. The HexaPro (F817P, A892P, A899P, A942P, K986P, and V987P) stabilizing mutations were introduced. The sequence was codon-optimized for mammalian host expression. Ancestral Wuhan HXP-S sequence is aligned with the new variant constructs: Beta, Gamma, and Delta. Changes in the N-terminal domain (NTD), the Receptor Binding Domain (RBD), and in the Spike 2 (S2) with respect to the original HXP-S sequence are highlighted in red, pink, and light purple, respectively. (B) NDV-HXP-S variants were rescued by reverse genetics as previously described (25). Cells were cotransfected with the expression plasmid required for replication and transcription of the NDV viral genome (NP, P, and L), together with the full length NDV cDNA. One day later, transfected cells were co-cultured with DF-1 cells. After 2 or 3 days, the tissue culture supernatants were inoculated into 8- or 9-day-old specific pathogen free (SPF) embryonated chicken eggs. Antigen identity was confirmed by biochemical methods and sequencing. The genetic stability of the recombinant viruses was evaluated across multiples passages on 10-day-old-SPF embryonated chicken eggs. NDV-HXP-S vaccine was inactivated with BPL and purified by sucrose cushion ultracentrifugation. (C) Comparison of NDV-HXP-S Delta virus versus NDV-HXP-S Delta with P681R mutation. The spike protein and NDV HN proteins were detected by Western blot using an anti-spike 2B3E5 mouse monoclonal antibody and an anti-HN 8H2 mouse monoclonal antibody, respectively. (D) Protein staining of NDV-HXP-S variant vaccines resolved on 4–20% SDS-PAGE. The viral proteins were visualized by Coomassie blue staining (L, S0, HN, N, P. and M). The uncleaved S0 spike protein is highlighted in blue with an approximate size of 200 kDa.