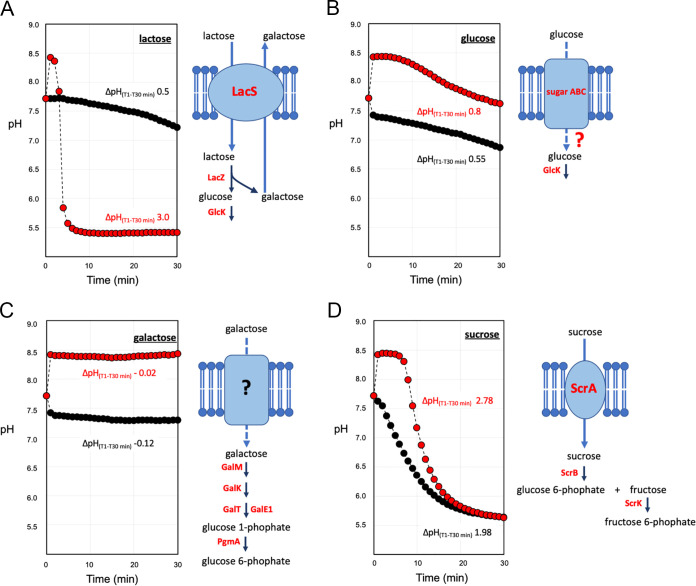
FIG 3.
Activation of homolactic fermentation in EdCs with different carbon sources. (A) EdCs activated with lactose. (B) EdCs activated with glucose. (C) EdCs activated with galactose. (D) EdCs activated with sucrose. EdCs activated with sugar and NH3 (red circles). EdCs activated with sugar (black circles). ΔpH(T1-T30 min) values are reported. Transport proteins and enzymes involved in sugar metabolism have also been reported. Transport proteins and enzymes detected in the proteome of EdCs are written in red. LacS, lactose permease. Sugar ABC, hypothetical glucose transporter. ScrA, sucrose PTS component II. LacZ, β-galactosidase. GlcK, glucose kinase. GalM, aldose 1-epimerase. GalK, galactokinase. GalT, galactose-1-phosphate uridylyltransferase. GalE1, UDP-glucose 4-epimerase. PgmA, phosphoglucomutase. Data are reported as the average of three replicates. The standard deviation was always below 2% of the measured pH.