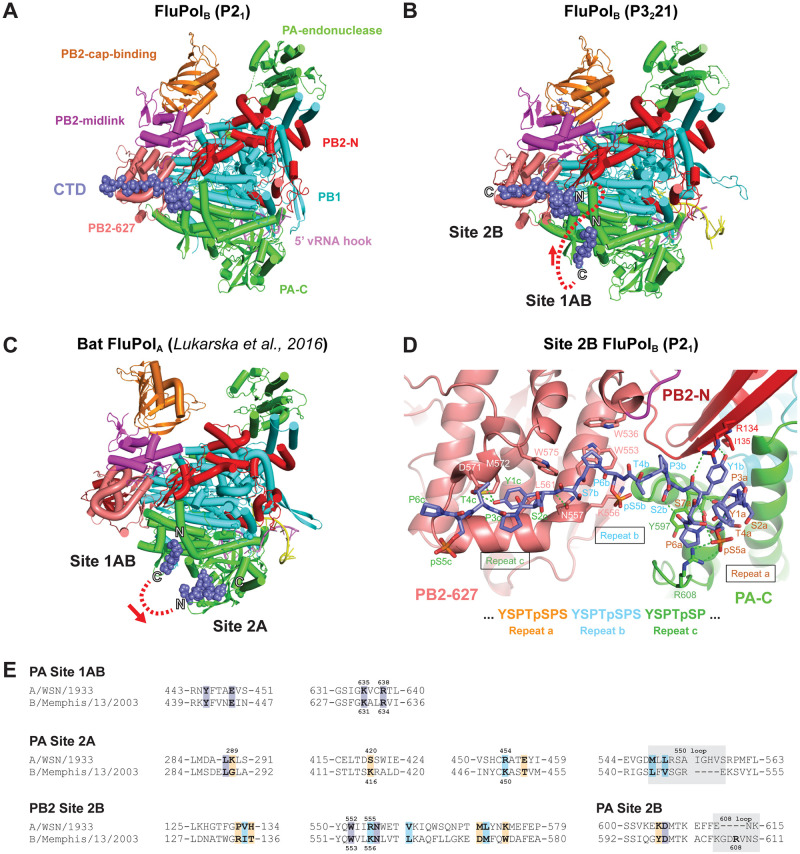
Fig 1. Structural analysis of CTD binding to influenza B polymerase.
A. Overall view of the crystal structure of influenza B polymerase, with bound vRNA 5’ hook (violet) and CTD peptide mimic (slate blue spheres) in site 2B. Ribbon diagram of the polymerase with PA (green), PB1 (cyan), PB2-N (red), PB2-cap-binding (orange), PB2-midlink (magenta), PB2-627 (deep salmon). B. Overall view of the crystal structure of influenza B polymerase with bound promoter (violet and yellow), capped primer (blue) and CTD peptide mimic (slate blue spheres) bound in sites 1AB and 2B. The polymerase is coloured as in (A). The N and C-termini of the two CTD fragments are marked and the red dotted line shows the shortest connection between them with directionality indicated by the arrow. C. Overall view of the crystal structure of bat influenza A polymerase with bound promoter and CTD peptide mimic (slate blue spheres) bound in sites 1AB and 2A ([32], PDB: 5M3H). The colour code is as in (A). The N and C-termini of the two CTD fragments are marked and the red dotted line shows the shortest connection between them with directionality indicated by the arrow. D. Details of the interaction between key residues of the influenza B polymerase PA subunit (green), PB2-N (red) and PB2-627 (deep-salmon) with the CTD peptide (slate blue sticks) in site 2B. Three CTD repeats denoted a (orange), b (cyan) and c (dark green) are involved in this interaction. Hydrogen bonds are indicated as dotted green lines. E. Sequence alignment of the CTD binding sites in the A/WSN/33 (A0A2Z5U3X0) and B/Memphis/13/2003 (Q5V8X3) polymerase subunits PA and PB2. Protein sequences were obtained from UniProt (https://www.uniprot.org/) and aligned with SnapGene 6.0. Key residues for CTD binding are indicated in bold. Identical, similar and non-similar residues are highlighted in purple, light blue and orange, respectively. Grey boxes indicate residues that form a loop. Residues submitted to mutagenesis in this study are indicated with their numbers above (FluPolA) and below (FluPolB) the alignment, respectively.