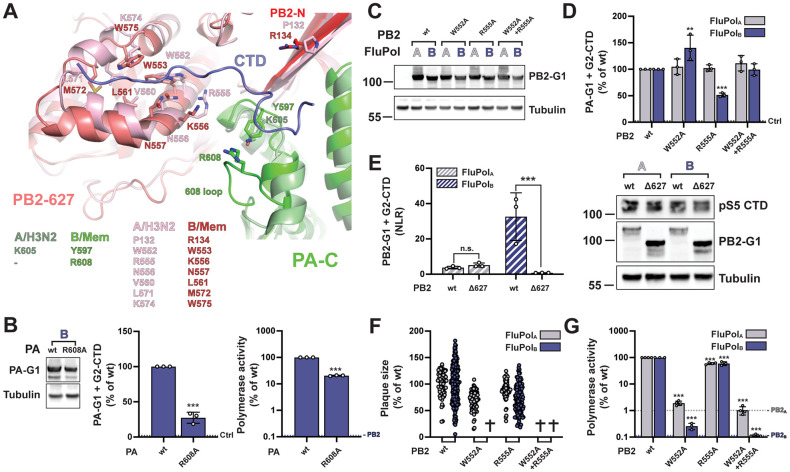
Fig 5. FluPolA and FluPolB CTD-binding mode at site 2B.
A. Superposition of CTD peptide (slate blue tube) bound in site 2B of FluPolB (B/Memphis/13/2003, PA green, PB2-N red, PB2-627 deep salmon) with the equivalent region of FluPolA (A/NT/60/1968 (H3N2), [58], PDB: 6RR7, PA light green, PB2 pink), showing similarities and differences in CTD interacting residues. See sequence alignment in Fig 1E. B. Protein expression (left), in vivo CTD binding (middle), and polymerase activity (right) of FluPolB PA R608A. Experiments were performed as described in Fig 3. The data shown are mean ± SD of three independent experiments performed in technical triplicates. ***p ≤ 0.001 (unpaired t test).C-D. Protein expression (C) and in vivo CTD binding (D) of CTD-binding site 2B mutants. Experiments were performed as described in Fig 3B and 3C for FluPolB and FluPolA site 1AB mutations. D. **p ≤ 0.002, ***p ≤ 0.001 (two-way ANOVA; Dunnett’s multiple comparisons test). E. In vivo CTD binding of PB2 Δ627 domain deletion mutants was investigated. G2-CTD was expressed by transient transfection in HEK-293T cells together with PB2-G1, PB1 and PA of FluPolA (A/WSN/1933, grey bars) or FluPolB (B/Memphis/13/2003, blue bars). Luciferase activities were measured in cell lysates at 24 hpt. Normalised luciferase ratios (NLRs) were calculated as described in the Materials and Methods section. ***p ≤ 0.001 (two-way ANOVA; Sidak’s multiple comparisons test). Cell lysates were analysed in parallel by western blot with antibodies specific for the pS5 CTD, G. princeps luciferase (PB2-G1) and tubulin. F-G. Characterisation of recombinant IAV and IBV viruses (F) and polymerase activity (G) of CTD-binding site 2B mutants. Experiments were performed as described in Fig 3D-E for FluPolB and FluPolA site 1AB mutations. F. (✞) no viral rescue. G. ***p ≤ 0.001 (two-way ANOVA; Dunnett’s multiple comparisons test).