Abstract
Acute kidney injury (AKI) is caused by renal ischemia/reperfusion injury (IRI) during kidney transplantation. The levels of both circular RNAs (circRNAs) and microRNAs (miRNAs/miR) appear to be critical for AKI detection. While several RNA interactions in AKI have been found, the regulatory mechanisms between the molecules remain to be fully elucidated. In the present study, miRNA expression profiling analysis was conducted using an online dataset to identify the differentially expressed miRNAs in rats with IRI. miR-328-3p was also found to be downregulated in human kidney-2 (HK-2) cells subjected to hypoxia/reperfusion (H/R), and its overexpression targeting pim-1 proto-oncogene (PIM1) resulted in an increased viability and a reduced apoptosis, as well as in the decreased expression of inflammatory factors upon H/R exposure. Putative targets and circRNAs of miR-328-3p were identified using publically available databases. The inhibition of circRNA integrin beta 1 (ITGB1; circITGB1) suppressed the inflammatory response induced by H/R by sponging miR-328-3p in HK-2 cells. Furthermore, a sequence of the functional ITGB1 promoter was studied for transcription factor GATA binding protein 1 (GATA1) binding sites. GATA1 binds to the ITGB1 promoter, leading to the expression of circITGB1. On the whole, the findings of the present study revealed a regulatory pathway modulating miR-328-3p in IRI, demonstrating that the GATA1-mediated regulation of circITGB1 enhanced the H/R-induced inflammatory response via the miR-328-3p/PIM1 axis.
Key words: acute kidney injury, circular RNA, microRNA, transcription factor, ITGB1, PIM1
Introduction
Kidney transplantation, a common treatment for end-stage renal disease, is one of the most common renal replacement therapies (1). Abnormal physiological conditions, such as the inflammation induced by vascular anastomosis or recanalization during transplantation, cause renal ischemia/reperfusion injury (IRI) or acute kidney injury (AKI) (2). IRI is an inevitable issue during organ transplantation, and it affects the therapeutic effects of kidney transplantation in relation to delayed graft functions and allograft dysfunction (3,4). Although previous studies have initially focused on prevention and treatment strategies for AKI, ~2% of hospitalized patients are diagnosed with AKI with a mortality rate of 12.4% (5). Hence, it is fundamental to understand the pathogenesis and explore novel treatment strategies for AKI.
MicroRNAs (miRNAs/miRs) are critical regulators of their target mRNAs by complementarily binding to mediate or transcript repression (6). miRNAs have recently been implicated in human renal diseases. In cancer cells, PinX1 has been found to activate cancer-related miR-125-3p, to cooperate with the VEGF signaling pathway and inhibit angiogenesis in renal cell carcinoma (7). miRNAs are also crucial diagnostic biomarkers for renal diseases and the interaction between miRNAs and downstream target genes results in targeted therapies for renal fibrosis (8) and AKI (9). For instance, the expression of miR-21 has been shown to be upregulated by kidney IRI in vitro and to contribute to tubular epithelial cell apoptosis and local inflammation (10). He et al (11) demonstrated that the expression of miR-328 was significantly decreased in TGF-β1-induced renal fibrogenesis and the overexpression of miR-328 decreased its target gene transcription and the cell epithelial-mesenchymal transition process. However, neither the effect of miR-328 on inflammatory response, nor the interaction with other non-coding RNAs to modulate mRNA transcription in AKI or IRI have been investigated to date, at least to the best of our knowledge.
CircularRNAs (circRNAs) are endogenous non-coding RNAs that are involved in the interaction between miRNAs and mRNAs in the 3′untranslated region (3′UTR) region (12). With advancements being made in next-generation sequencing technology and bioinformatics, specific circRNA signatures have been identified in several human diseases (13). In a previous study, the absence of hsa_circ_0068888 in a cell model of lipopolysaccharide-induced AKI suggested that the overexpression of circ_0068888 may be involved in the inflammatory response and oxidative stress via sponging miR-21-5p (14). Kölling et al (15) found that circRNA-126 expression in patients with AKI was a putative predictor of the survival rate. However, there are only a few circRNAs whose functions have been fully elucidated to date.
The present study analyzed RNA-sequencing data and identified a cohort of miRNAs that are aberrantly expressed in AKI in a rat model. miR-328-3p was further validated as a downregulated miRNA in a cell model of AKI; miR-328-3p expression was decreased by circRNA integrin beta 1 (circITGB1, circbase ID: hsa_circ_0018148). In addition, experiments performed for the silencing of circITGB1 and miR-328-3p in human kidney-2 (HK-2) cells subjected to hypoxia/reperfusion indicated that the silencing of circITGB1 upregulated the expression level of miR-328-3p and decreased Pim-1 proto-oncogene (PIM1) expression. The downregulation of circITGB1 exerted an anti-inflammatory effect in AKI. Moreover, it was observed that the gene symbol of circITGB1, ITGB1, was subjected to regulation by the transcription factor GATA binding protein 1 (GATA1). Hence, therapeutic approaches aimed at targeting GATA1/circITGB1/miR-328-3p/PIM1 may prove to be beneficial for AKI prevention and therapy.
Materials and methods
Cell culture and H/R treatment
The HK-2 cell line (CRL-2190, ATCC) was grown in complete growth medium, keratinocyte serum-free medium (K-SFM, 17005-042, Gibco; Thermo Fisher Scientific, Inc.) supplemented with 0.05 mg/ml bovine pituitary extract and 5 ng/ml epidermal growth factor. The HK-2 cells were incubated at 37°C in a suitable incubator containing a 5% CO2 in air atmosphere. To induce H/R, the HK-2 cells were treated with K-SFM medium and subjected to 12 h of hypoxia (5% CO2, 1% O2 and 94% N2), followed by cultivation in complete medium for 0/3/6/9/12 h of reoxygenation (5% CO2, 21% O2 and 74% N2).
Reverse transcription-quantitative (RT-qPCR)
The expression of mRNAs, circRNAs and miRNAs was analyzed following the cell treatment or transfection. Total RNA was isolated from the HK-2 cells using Trizol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.). Total RNA (1.5 μg) was used for cDNA synthesis at 42°C with SuperScript™ IV revertase (Invitrogen; Thermo Fisher Scientific, Inc.). qPCR reactions were performed with SYBR®-Green Mix (Applied Biosystems; Thermo Fisher Scientific, Inc.) for mRNA and circRNA expression analysis, with the TaqMan™ MicroRNA Assay kit (Applied Biosystems; Thermo Fisher Scientific, Inc.) for miRNA expression analysis. The specific primer pairs of ITGB1 (NCBI Reference Sequence NM_002211), PIM1 (NM_002648), neurofibromin 2 (merlin) (NF-2; NM_181833), GAPDH (NM_001357943), GATA1 (NM_0020nf-49) and ubiquitin protein ligase E3 component n-recognin 4 (UBR4; NM_020765.3) were designed using NCBI Primer-BLAST (https://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome). The stem-loop primer of miR-328-3p (miRbase ID: MIMAT0000752) was designed using the Primer3 (https://primer3.ut.ee/) online tool. In addition, Primer3 was utilized to design the divergent primers of circITGB1 (CircBank ID hsa_circ_0018148), circARL6IP1 (CircBank ID hsa_circ_0038229) and circH-NRNPF (CircBank ID hsa_circ_0018263). After obtaining the divergent primer sets as aforementioned, CircPrimer 2.0 software (downloaded from https://www.bio-inf.cn/; DOI: 10.1186/s12859-018-2304-1) was utilized to examine the specificity of the circRNA primers. The lists of primers for each mRNA, circRNA and miRNA are presented in Table I. Relative expression was analyzed using the 2−ΔΔCq method (16) and normalization with internal controls (GAPDH or U6). As for RNase R treatment, total RNA (2 μg per group) of HK-2 cells were incubated with RNase R (a final concentration of 10 μg/ml) for 3 min at 37°C (Beijing Solarbio Science & Technology Co., Ltd.). RT-qPCR was performed to determine the expression levels of ITGB1 mRNA and circITGB1. To identify the circular feature of circITGB1, Oligo(dT) 18 primers (Beijing Solarbio Science & Technology Co., Ltd.) and random primers (Beijing Solarbio Science & Technology Co., Ltd.) were employed to perform cDNA synthesis, respectively. The resulting cDNAs were then detected by qPCR using the primers targeting circITGB1 or ITGB1, respectively.
Table I.
Sequences of primers used in the present study.
| Gene | Primer sequence (5′→3′) |
|---|---|
| hsa_circ_0038229-F | TTTGTTTTGTGTAAAAGGGGAAA |
| hsa_circ_0038229-R | GGTTGGTGCTGCGATTATCT |
| hsa_circ_0018263-F | CACATCTGTTGATAGCTGGAGAA |
| hsa_circ_0018263-R | GAGCTAAGGCTACGGGAACC |
| circITGB1-F | GGATGTTGACGACTGTTGGTT |
| circITGB1-R | CAATTTGGCCCTGCTTGTAT |
| UBR4-F | TGTCAACAATGGCAACCCCT |
| UBR4-R | GCTGATGCTGTGGTGTCAGA |
| ITGB1-F | GACGCCGCGCGGAAAA |
| ITGB1-R | ACATCGTGCAGAAGTAGGCA |
| PIM1-F | CCCGACAGTTTCGTCCTGAT |
| PIM1-R | ACCCGAAGTCGATGAGCTTG |
| NF-2-F | GGGCTAAAGGGCTCAGAGTG |
| NF-2-R | GAGTCCGGCACACCAAATCA |
| GATA1-F | GGACAGGCCACTACCTATGC |
| GATA1-R | CTTTTCCAGATGCCTTGCGG |
| GAPDH-F | CTGGGCTACACTGAGCACC |
| GAPDH-R | AAGTGGTCGTTGAGGGCAATG |
| miR-328-3p-RT | CTCAACTGGTGTCGTGGAGTCG |
| GCAATTCAGTTGAGACGGAAGG | |
| miR-328-3p-F | ACACTCCAGCTGGGCTGGCCCT |
| CTCTGCCC | |
| U6-RT | GTCGTATCCAGTGCGTGTCGTG |
| GAGTCGGCAATTGCACTGGAT | |
| ACGACAAAATATGGAAC | |
| U6-F | TGCGGGTGCTCGCTTCGGCAGC |
| Universal reverse | TATCC AGTGCGTGTCGTG |
| ITGB1 promoter-F | GCACGGACCAATTATGCTTT |
| ITGB1 promoter-R | CTAGGAGGAGGCGGAGGAT |
| ITGB1 promoter site1-F | AGACTGGGCC AATTCTGATG |
| ITGB1 promoter site1-R | GCCAATCTGAAGACC TCTAGGA |
| ITGB1 promoter site2-F | TCAAGTTTTGATTTTGCTGGT |
| ITGB1 promoter site2-R | GGACTGGAATGCCC ATTTG |
| ITGB1 promoter site3-F | GGGCATTCC AGTCCC TAATC |
| ITGB1 promoter site3-R | GGCAAGCTGCTATCC TGGTA |
| ITGB1 promoter site4-F | CAATCATCGAATCGACATGC |
| ITGB1 promoter site4-R | AGTTGCGTCC TGCTTTTGAT |
F, forward; R, reverse; ITGB1, integrin beta 1; PIM1, pim-1 proto-oncogene; GATA1, GATA binding protein 1; UBR4, ubiquitin protein ligase E3 component n-recognin 4; NF-2, neurofibromin 2.
Cell transfection
The knockdown of circITGB1, PIM1 and GATA1 was achieved by transfecting the HK-2 cells with small interfering RNAs (siRNAs). siRNAs targeting PIM1 (NM_002648), circITGB1 (circbase ID: hsa_circ_0018148) and GATA1 (NM_002049) were designated as siPIM1, si-circITGB1 and si-GATA1, respectively. siPIM1 and si-GATA1 was designed using BLOCK-iT™ RNAi Online Designer (http://rnaidesigner.thermofisher.com/rnaiex-press/design.do). In addition, the Circinteractome online database (https://circinteractome.irp.nia.nih.gov/index.html) (17) was utilized to design the siRNA specific to circITGB1. All siRNA sequences were synthesized by Shanghai Sango Biotechnology Co., Ltd. and were as follows: siPIM1 sense, 5′-CGGGUUUCUCCGGCGUCAUUAGGdTdT-3′ and antisense, 5′-CCUAAUGACGCCGGAGAAACCCGdTdT-3′; si-circITGB1 sense, 5′-GUGGAGAAUCCAGAUGAAAAUdTdT-3′ and antisense, 5′-AUUUUCAUCUGGAUUCUCCACdTd T-3′; si-GATA1 sense, 5′-CCCAGUCUUUCAGGUGUACCCAUUGdTdT-3′ and antisense, 5′-CAAUGGGUACACCUGAAAGACUGGGdTdT-3′. The miR-328-3p mimics, inhibitors and negative controls were obtained from Guangzhou RiboBio Co., Ltd. and the sequences were as follows: miR-328-3p mimic (miR10000752-1-5) sense, 5′-CUGGCCCUCUCUGCCCUUCCGU-3′ and antisense, 5′-GGAAGGGCAGAGAGGGCCAGUU-3′; NC mimic (miR1N0000001-1-5) sense, 5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense, 5′-ACGUGAACAGUUCGGAGAATT-3′; miR-328-3p inhibitor (miR20000752-1-5) sense, 5′-ACGGAAGGGCAGAGAGGGCCAG-3′; NC inhibitor (miR2N0000001-1-5), 5′-CAGUACUUUUGUGUAGUACAA-3′. The GATA1 coding sequence fragment (~1.4 kb) was amplified with a primer set (forward, 5′-ACTGAGCTTGCCACATCCCC-3′ and reverse, 5′-GGGCCCAGAGACTTGGGTTG-3′) and inserted into the pcDNA3.1 vector (Invitrogen; Thermo Fisher Scientific, Inc.) between the BamH1 and XhoI sites. The plasmid construct was transformed in E. coli, and selected colonies were then cultured. Plasmid DNA was purified using the Plasmid Maxi Preparation kit (Beyotime Institute of Biotechnology). To perform cell transfection, the HK-2 cells were plated in 96-well plates and incubated for 24 h at 37°C. Each 1 μg of plasmids (or 50 nM miRNA mimic or 100 nM miRNA inhibitor) were transfected into the HK-2 cells using Lipofectamine™ 3000® Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h, the HK-2 cells were subjected to H/R (12 h of hypoxia and 6 h of reoxygenation). Additionally, the transfection efficiency was examined using RT-qPCR.
Detection of cell viability
The HK-2 cells were transfected with the siRNAs or miRNA mimic or miRNA inhibitor for 48 h as described above and then subjected to H/R. The cells were then collected and plated in a 96-well plate (1,000 cells/well). Following cell adherence, Cell Counting Kit-8 (CCK-8) solutions (Beijing Solarbio Science & Technology Co., Ltd.) were added to the cells for detecting cell viability. At 2 h following the addition of CCK-8, the absorbance at 450 nm was measured using a microplate reader (DNM-9606, Beijing Pulangxin technology Co., Ltd.) and cell the viability rate was analyzed compared to the control group (100%).
Detection of cell apoptosis
The HK-2 cells were subjected to H/R as described above. For cell apoptosis analysis, 1×106 cells were harvested using trypsase solution (Beijing Solarbio Science & Technology Co., Ltd.) and resuspended in binding buffer (Beijing Solarbio Science & Technology Co., Ltd.). Following centrifugation, the pellets were incubated with 5 μl Annexin V-Fluorescein isothiocyanate isomer (FITC) solutions (Beijing Solarbio Science & Technology Co., Ltd.) for 10 min and then with with 5 μl propidium iodide (PI) solutions (Beijing Solarbio Science & Technology Co., Ltd.). Flow cytometry was performed using a Guava EasyCyte Mini System (Luminex, Inc.).
Detection of inflammatory cytokines
Enzyme linked immunosorbent assay (ELISA) interleukin-1β (IL-1β), interleukin-6 (IL-6) and tumor necrosis factor-α (TNF-α) kits (Beijing Solarbio Science & Technology Co., Ltd.) were utilized for the quantitative determination of concentrations in the cell culture supernatants. Firstly, 100 μl samples were added to each well and incubated for 90 min at 37°C. The cells were then incubated with 100 μl working solution (a component of the ELISA kits) containing biotin-conjugated anti-human IL-1β/IL-6/TNF-α antibodies (a component of the ELISA kits) for 60 min at 37°C. Subsequently, 100 μl streptavidin-horseradish peroxidase (HRP) working solutions (a component of ELISA kit) were utilized for a further 30 min of incubation at 37°C. Following incubation with 100 μl substrate solution (a component of the ELISA kits) away from light at 37°C for 15 min, the reactions were terminated using stop solutions (a component of the ELISA kits) and the absorbance at 450 nm was measured using a microplate reader (DNM-9606, Beijing Pulangxin technology Co., Ltd.).
RNA-sequencing analysis
Using the keywords 'renal ischemia reperfusion' and 'microRNA', datasets were searched in the GEO database (https://www.ncbi.nlm.nih.gov/gds/). The selection criteria were as follows: i) Datasets with non-coding RNA profiling types; ii) datasets containing a sham operation group and a renal IRI group; iii) datasets with reasonable biological replications. The GSE124669 dataset contains samples from multiple time points post-IRI. The GSE124669 dataset was used for RNA-seq analysis. In brief, Sonoda et al (18) constructed a bilateral IRI model. Sprague-Dawley rats (male, 10 weeks) were randomly divided into the sham operation group and IRI group. The left and right renal vascular pedicles were subjected to clamped operation for 25 min and then reperfused with blood. The RNA used for sequencing was isolated from urinary exosomes at days 1, 2, 3, 7 and 14 post-IRI. The RNA samples were then sequenced using the Illumina HiSeq 2500 platform. Raw sequence files were obtained from the SRA (SRP175286, https://www.ncbi.nlm.nih.gov/sra). The RNA sequencing data were obtained from Gene Expression Omnibus (GSE124669). The reads containing adapter sequences and low-quality reads (Phred-scale quality score <20) were removed from the raw data. The sequence quality was assessed using FastQC (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/, released Version 0.11.9). Clean data were subjected to analysis using the IDEG6.2 online tool (http://telethon.bio.unipd.it/bioinfo/IDEG6_form/) (19). The differentially expressed miRNAs with ±1.5-fold change (FC) and an adjusted P-value <0.05 were obtained using RNA sequencing analysis. The differentially expressed miRNA (rno-miR-328a-3p) was then subjected to an analysis on the UCSC Genome Browser (https://blast.ncbi.nlm.nih.gov/Blast.cgi) to confirm the conservation between rno-miR-328a-3p and hsa-miR-328-3p.
Prediction of miRNA-mRNA interactions
The mRNAs targeting miR-328-3p were predicted using three different databases: TargetScan (http://www.targetscan.org/vert_72/), Pictar (https://pictar.mdc-berlin.de/cgi-bin/new_PicTar_mouse.cgi) and miRDB (http://mirdb.org/). The overlapping interactions were calculated utilizing a web tool (http://bioinformatics.psb.ugent.be/webtools/Venn/). A graphical output in the form of venn diagram was produced to illustrate the overlapping mRNA interactions.
Prediction of miRNA-circRNA interactions
The binding sites between circRNAs and miR-328-3p were predicted using StarBase v2.0 (http://starbase.sysu.edu.cn/). StarBase perform miRNA-circRNA interactions by intersecting the predicting target sites of miRNAs with binding sites of Ago protein, which were derived from CLIP-seq data. The Ago clipExpNum is defined as the number of supporting AGO CLIP-seq experiments. An additional screening parameter (8mer class, clipExpNum strict stringency ≥5) was changed to refine the results. The 51 candidate circRNAs of miR-328-3p are listed in Table II. The 51 candidate circRNAs were sorted using Ago clipExpNum in descending numerical order. The top three circRNAs with a high clipExpNum were subjected to further RT-qPCR analysis in the cells subjected to H/R.
Table II.
Candidate target circRNAs of miR-328-3p.
| AgoClip ExpNum | CircRNA | Gene symbol | AgoClip ExpNum | CircRNA | Gene symbol |
|---|---|---|---|---|---|
| 33 | hsa_circ_0038229 | ARL6IP1 | 7 | hsa_circ_0060786 | ZNFX1 |
| 24 | hsa_circ_0018263 | HNRNPF | 6 | hsa_circ_0011360 | TXLNA |
| 20 | hsa_circ_0018148 | ITGB1 | 6 | hsa_circ_0011535 | ZMYM4 |
| 17 | hsa_circ_0067884 | PHC3 | 6 | hsa_circ_000837 | ZMYM4 |
| 15 | hsa_circ_0038615 | UBFD1 | 6 | hsa_circ_0015760 | ASPM |
| 15 | hsa_circ_0060840 | ADNP | 6 | hsa_circ_0064170 | ARPC4 |
| 15 | hsa_circ_0092035 | RPL10 | 6 | hsa_circ_0064168 | ARPC4 |
| 14 | hsa_circ_0022454 | MTA2 | 6 | hsa_circ_0019002 | WAPAL |
| 11 | hsa_circ_0052438 | PXDN | 6 | hsa_circ_0024149 | CASP4 |
| 11 | hsa_circ_0032208 | ZBTB25 | 6 | hsa_circ_0035341 | ARPP19 |
| 10 | hsa_circ_0076407 | KIAA0240 | 6 | hsa_circ_0042407 | AKAP10 |
| 10 | hsa_circ_0017865 | VIM | 6 | hsa_circ_0044502 C | OL1A1 |
| 9 | hsa_circ_0070224 | HNRPDL | 6 | hsa_circ_0051287 | PAFAH1B3 |
| 9 | hsa_circ_0070499 | ADH5 | 5 | hsa_circ_0012093 | PTPRF |
| 9 | hsa_circ_0002819 | NEMF | 5 | hsa_circ_0016274 | YOD1 |
| 8 | hsa_circ_0008494 | ARID1A | 5 | hsa_circ_0058542 | TRIP12 |
| 8 | hsa_circ_0081562 | SERPINE1 | 5 | hsa_circ_0074380 | NR3C1 |
| 8 | hsa_circ_0026833 | ESYT1 | 5 | hsa_circ_0074379 | NR3C1 |
| 8 | hsa_circ_0026859 | ESYT1 | 5 | hsa_circ_000420 | NR3C1 |
| 8 | hsa_circ_0041373 | HIC1 | 5 | hsa_circ_0085911 | PLEC |
| 8 | hsa_circ_0090446 | TIMP1 | 5 | hsa_circ_0085897 | PLEC |
| 7 | hsa_circ_0015610 | SMG7 | 5 | hsa_circ_0089684 | C9orf167 |
| 7 | hsa_circ_0015612 | SMG7 | 5 | hsa_circ_0025304 | PHB2 |
| 7 | hsa_circ_0007774 | EDAR | 5 | hsa_circ_0036012 | SMAD3 |
| 7 | hsa_circ_0075320 | SQSTM1 | 5 | hsa_circ_0048442 | AES |
| 7 | hsa_circ_0006271 | GIGYF1 |
AgoClip ExpNum, number of supporting AGO CLIP-seq experiments.
RNA immunoprecipitation (RIP) assay
RIP assay was carried out to confirm the RNA-Ago2 protein interaction by utilizing the Dynabeads™ Protein A Immunoprecipitation kit (Invitrogen; Thermo Fisher Scientific, Inc.). Briefly, the HK-2 cells overexpressing circITGB1 or miR-328-3p were incubated in a 25 cm2 cell culture flask for 48 h. A total of 5×106 cells were lysed using cell extraction buffer. Dynabeads magnetic beads were resuspended on a roller for 5 min. Subsequently, 50 μl (1.5 mg) beads were placed on the magnet to separate the beads from the solution. Typically, 1 μg Anti-Ago2 (ab186733, Abcam) or IgG (ab172730) antibodies diluted in 200 μl Ab Binding Buffer were incubated with the beads for 10 min at room temperature. The tube was placed on the magnet to remove the supernatant. The magnetic bead-Ab complex was resuspended in 200 μl Ab Binding and washing buffer (a component of the immunoprecipitation kit). To allow the antigen (Ag) to bind to the bead-Ab complex, the cell sample (100 μl) containing RNAs were incubated with the bead-Ab complex in a shaker for 10 min. The tube containing the bead-Ab-Ag complex was placed on the magnet to remove the supernatant. The bead-Ab-Ag complex were washed with 200 μl washing buffer three times. The bead-Ab-Ag complex was then separated on the magnet between each wash. The resulting complex was resuspended in 100 μl washing buffer and was transferred to a clean tube. The tube was placed on the magnet to remove the supernatant. A total of 20 μl elution buffer (a component of the immunoprecipitation kit) was then incubated with the beads-Ab-Ag complex for 2 min to dissociate the complex. The tube was placed on the magnet. The supernatant containing eluted Ab and Ag was transferred to a clean tube. The coprecipitated RNAs were isolated from the supernatant by using RNeasy MinElute Cleanup kit (Qiagen, Inc.) and reverse transcribed into cDNA using the Prime-Script RT reagent kit (Takara Biotechnology Co., Ltd.) according to the manufacturer's instructions. Subsequently, the mRNA levels of circITGB1 and miR-328-3p were measured using RT-qPCR assay as described above.
Transcription factor analysis
The in silico analysis of the ITGB1 promoter (2,000 bp upstream of the transcriptional start site) was performed to identify the binding sites for GATA1. The sequence of the ITGB1 promoter was downloaded from NCBI (https://www.ncbi.nlm.nih.gov/). The JASPAR database (http://jaspar.genereg.net/) provided the frequency matrix of transcription factor GATA1 (Matrix ID: MA0035.4). A FASTA-formatted sequence (ITGB1 promoter) was input to scan with the GATA1 matrix model. A total of four putative sites were predicted with a relative profile score threshold of 80%.
Luciferase reporter gene assay
Four fragments in the ITGB1 promoter were amplified using PCR primers (Table I). The fragments of the ITGB1 promoter were inserted into the pGL3 luciferase vector (Promega Corporation) and the resulting vectors were named as ITGB1 promoter-luc or site1/2/3/4-WT, respectively. The QuickMutation™Plus Site-Directed Mutagenesis kit (Beyotime Institute of Biotechnology) was utilized to construct mutated ITGB1 promoter luciferase reporter vectors. Cells in which GATA1 was overexpressed or silenced (1×105) were seeded in 96-well plates and co-transfected with the luciferase plasmid (2 μl/μg) using Lipofectamine™ 3000® (Invitrogen; Thermo Fisher Scientific, Inc.). After 48 h, a luciferase reporter assay kit (Promega Corporation) was utilized for the detection of the luciferase activities. The relative luciferase activities were analyzed via normalization against the Renilla luciferase values.
Western blot analysis
Cells were collected by incubation with trypsin and lysed with radio immunoprecipitation assay (RIPA) buffer (Beijing Solarbio Science & Technology Co., Ltd.) for 30 min. Following centrifugation at 800 × g for 5 min at 4°C (Beckman Coulter Life Sciences), the concentration of total protein were determined using BCA Protein Assay kit (Beijing Solarbio Science & Technology Co., Ltd.). The proteins were then denatured at 95°C and then subjected to separation on 12% SDS-PAGE gels. A total of 20 μg proteins were then transferred onto polyvinylidene fluoride (PVDF) membranes and blocked with bovine serum albumin (BSA) buffer for 2 h at room temperature. The membranes were incubated with primary antibodies that were diluted in BSA buffer overnight at 4°C. The primary antibodies used were anti-PIM1 (1:2,000, ab54503), anti-NF-2 (1:10,000, ab109244) and anti-GAPDH (1:1,000, ab8245) (all from Abcam). The PVDF membranes were then incubated with HRP-conjugated secondary antibodies (1:1,000, ab7090; 1:5,000, ab97080, Abcam) for 2 h at room temperature. Finally, immunoreactive proteins were detected using enhanced chemiluminescence (ECL) Western Substrate (Thermo Fisher Scientific, Inc.) and visualized using a Chemiluminescence Imager (Tanon Science & Technology).
Statistical analysis
Data are presented as the mean ± standard deviation (SD) from three independent experiments. Statistical analysis between multiple groups was performed with one-way analysis of variance (ANOVA) and Bonferroni's post hoc comparisons test using GraphPad Prism 8.0 software (GraphPad, Inc.). A P-value <0.05 was considered to indicate a statistically significant difference.
Results
miR-328-3p is downregulated in HK-2 cells subjected to H/R
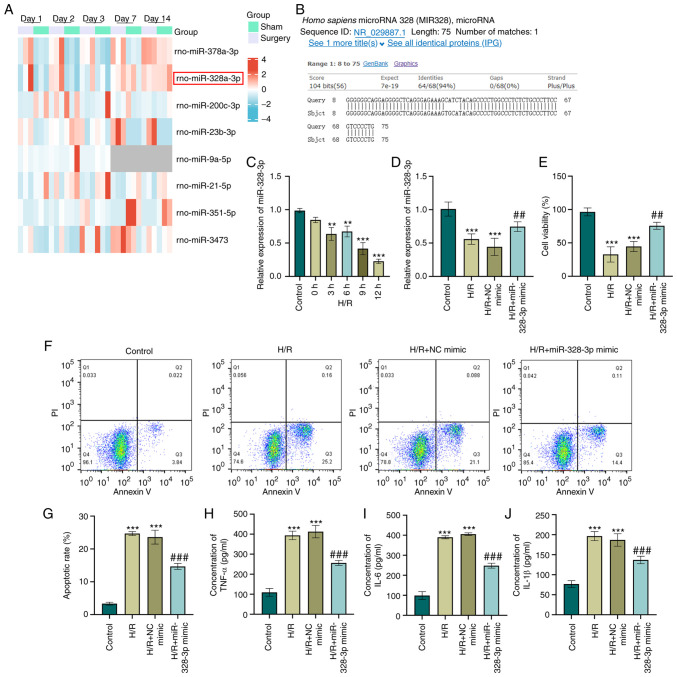
miRNAs are one of the most genetically expressed non-coding RNAs in AKI (20) and consequently potentiate or inhibit the pathogenicity of inflammation (21). With an objective to explore the miRNAs regulated in AKI, a RNA sequencing dataset (GSE124669) was obtained and data analysis was performed. miRNA expression profiles in rats were clustered based on the treatment conditions (IR surgery group and sham group). The statistical analysis of the RNA-sequencing data with ±1.5 FC and adjusted P-value <0.05 generated a list that contained 36 miRNAs that were differentially expressed in the IR surgery groups at each time points compared with the sham groups. The heatmap displayed a total of eight miRNAs (rno-miR-378a-3p, rno-miR-328a-3p, rno-miR-200c-3p, rno-miR-23b-3p, rno-miR-9a-5p, rno-miR-21-5p, rno-miR-351-5p and rno-miR-3473) that were differentially expressed at two time points (Fig. 1A). It was previously reported that miR-378a-3p plays a crucial role in ferroptosis in AKI (22). The present study focused on the second highest differentially expressed miRNA (rno-miR-328a-3p). rno-miR-328a-3p was downregulated in the IR group on days 1/2/3. To explore the potential function of rno-miR-328a-3p in cell development and regulation, a conservation analysis we performed using UCSC Genome Browser (https://blast.ncbi.nlm.nih.gov/Blast.cgi). The results indicated that the precursor sequence for both rat rno-miR-328a-3p and human hsa-miR-328-3p was highly conserved with a 94% sequence identity and only a four nucleotide difference (Fig. 1B). In order to elucidate whether miR-328-3p expression has any effect on cells damaged by H/R, HK-2 cells were subjected to hypoxia for 12 h and then cultured in a normoxic incubator for 0/3/6/9/12 h. The validation of miR-328-3p expression using RT-qPCR analysis was performed and the results revealed a reoxygenation-dependent inhibition of miR-328-3p levels in HK-2 cells (P<0.01 and P<0.001, Fig. 1C). The 9-h and 12-h reoxygenation decreased the survival rate of the HK-2 cells by ~90%. The cell amount was not sufficient to perform subsequent experiments. As the 9-h and 12-h reoxygenation may exert cytotoxic effects on HK-2 cells, the 6-h reoxygenation time point was selected for use in further experiments. Thus, it was concluded that miR-328-3p downregulation was induced by H/R in HK-2 cells.
Figure 1.
miR-328-3p is downregulated and exerts anti-inflammatory effects in H/R-exposed HK-2 cells. (A) Hierarchical clustering of the expression array data (GSE124669) in rats subjected to ischemia/reperfusion injury and sham-operated rat. (B) Sequence alignment between rno-miR-328a-3p and human hsa-miR-328-3p. (C) HK-2 cells were subjected to hypoxia (12 h) and reoxygenation for various periods of time (0, 3, 6, 9 and 12 h). The expression level of miR-328-3p was then determined using RT-qPCR. (D) Expression levels of miR-328-3p in HK-2 transduced with NC mimic or miR-328-3p mimic and exposed to with H/R were examined using RT-qPCR analysis. (E) The miR-328-3p-overexpressing cells or NC mimic-transfected cells were seeded in a 96-well culture dish and exposed to H/R (12 h of hypoxia and 6 h of reoxygenation) and evaluated for cell viability using CCK-8 assay by detecting the absorbance at 450 nm. (F and G) The miR-328-3p-overexpressing or NC mimic-transfected cells were exposed to H/R to stimulate acute kidney injury, followed by staining with Annexin V or PI solutions, and examined using flow cytometry. (H-J) ELISA of TNF-α, IL-6 and IL-1β pro-inflammatory markers in HK-2 cells following transfection with miR-328-3p mimic/NC mimic and treated with H/R. Data are presented as the mean ± standard deviation from three independent experiments. **P<0.01 and ***P<0.001 vs. control group; ##P<0.01 and ###P<0.001 vs. H/R + NC mimic group. RT-qPCR, reverse transcription-quantitative PCR; H/R, hypoxia/reperfusion; IL-1β, interleukin-1β; IL-6, interleukin-6; TNF-α, tumor necrosis factor-α.
miR-328-3p exerts anti-inflammatory effects in HK2 cells subjected to H/R
Since miR-328-3p was expressed at low levels following H/R, it was overexpressed by miR-328-3p mimic transfection in HK-2 cells and its effects on various parameters related to renal injury were examined. The overexpression efficiency was confirmed using RT-qPCR assay (P<0.01, Fig. S1A). As shown in Fig. 1D, mimic transfection indeed led to a high level of miR-328-3p in the H/R-exposed cells compared with the H/R group (P<0.01 and P<0.001). It was observed that H/R induced a significant decrease in the viability of the HK-2 cells (P<0.001, Fig. 1E). Notably, the re-expression of miR-328-3p significantly increased cell viability when compared to H/R treatment alone (P<0.01, Fig. 1E). Moreover, while H/R treatment increased the cell apoptotic rate, the overexpression of miR-328-3p reversed the promoting effects of H/R on HK-2 cell apoptosis (P<0.001, Fig. 1F and G). The concentrations of inflammation-related cytokines were also significantly increased in the H/R group, as shown using ELISA (P<0.001, Fig. 1H-J). However, the overexpression of miR-328-3p inhibited the inflammatory response triggered by H/R (P<0.001, Fig. 1H-J). These results indicated that the pro-inflammatory effects of H/R on HK-2 cells were partly reversed by the overexpression of miR-328-3p.
miR-328-3p targets the PIM1 gene and abrogates the H/R-mediated inflammatory response in HK-2 cells
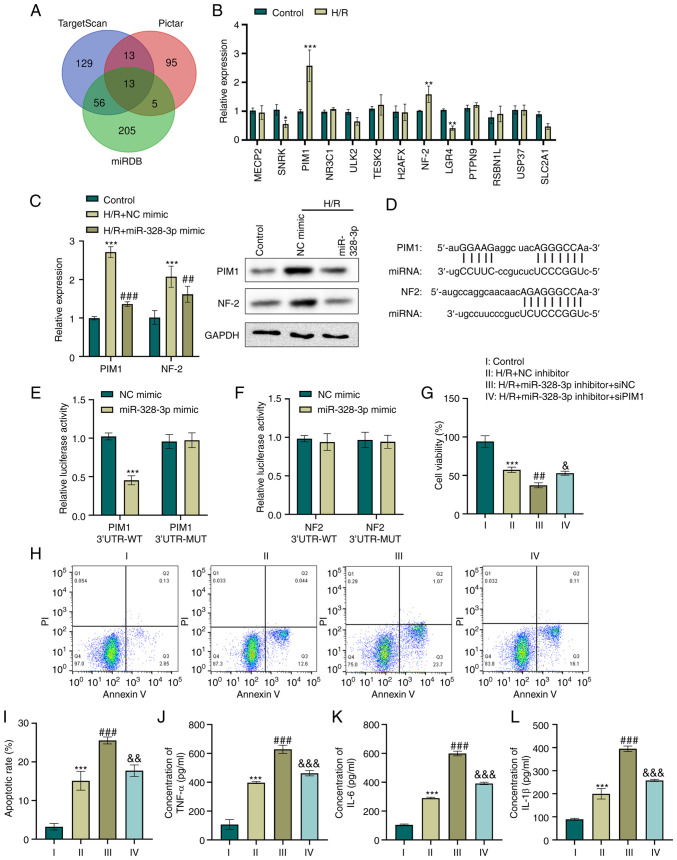
One of the mechanisms through which miRNAs function in cell regulation is competitively targeting the 3′UTR region of target genes, which leads to the loss or gain of gene function and controls cellular biological behaviors (23). In the present study, bioinformatics analysis integrated with some online databases predicted a total of 13 genes targeting miR-328-3p (Fig. 2A). Furthermore, the expression levels of the predicted targets were screened in HK-2 cells following H/R. The results revealed that the exposure of HK-2 cells to H/R resulted in a >1.5-fold increase in PIM1 and NF-2 levels compared to the control group (P<0.01 and P<0.001, Fig. 2B). Subsequently, the present study examined whether PIM1 or NF-2 expression was regulated by miR-328-3p in H/R-e HK-2 cells. H/R led to the increased expression of PIM1 and NF-2, as determined using RT-qPCR and western blot analysis (P<0.001, Fig. 2C). Moreover, PIM1 and NF-2 expression was considerably lower in the H/R + miR-328-3p mimic group than in the H/R group (P<0.01 and P<0.001, Fig. 2C). The scanning of miR-328-3p for human PIM1 and NF2 gene recognition sites, revealed an 8mer complementary binding site in the 3′UTR region of PIM1 mRNA and 7mer-8 sequence complementary to the 3′UTR region of NF2 mRNA, as shown in Fig. 2D. To elucidate whether miR-328-3p targets PIM1 and NF2, the putative miR-328-3p recognition sequences of targets were cloned in the pMIR reporter vector. Co-transfection with PIM1 3′UTR-WT and miR-328-3p mimic in HK-2 cells significantly reduced the luciferase activity (P<0.001, Fig. 2E). In addition, the mutant PIM1 sequence in the putative miR-328-3p binding sites abolished this effect (Fig. 2E). However, luciferase reporter gene assay indicated that miR-328-3p failed to target the NF-2 3′UTR (Fig. 2F).
Figure 2.
miR-328-3p targets the PIM1 gene and abrogates the H/R-mediated inflammatory response in HK-2 cells. (A) Venn diagram illustrating common targeting gene candidates of miR-328-3p between the TargetScan, Pictar and miRDB databases. (B) HK-2 cells were subjected to hypoxia (12 h) and reoxygenation for 6 h. The expression levels of gene candidates were then determined using RT-qPCR. *P<0.05, **P<0.01 and ***P<0.001 vs. control group. (C) The expression levels of PIM1 and NF-2 in HK-2 transduced with NC mimic or miR-328-3p mimic and treated with H/R were examined using RT-qPCR and western blot analysis. ***P<0.001 vs. control group; ##P<0.01 and ###P<0.001 vs. H/R + NC mimic group. (D) Sequence alignment between miR-328-3p seed sequence and 3′UTR sequences of targeting genes. (E and F) Luciferase reporter gene assays were carried out with miR-328-3p mimic or NC mimic transfection in cells co-transfected with PIM1 3′UTR-WT/MUT or NF-2 3′UTR-WT/MUT. ***P<0.001 vs. NC mimic group. HK-2 cells were co-transfected with miR-328-3p inhibitor/NC inhibitor and siPIM1/siNC and then treated with H/R. (G) Cell viability was evaluated using CCK-8 assay by detecting the absorbance at 450 nm. (H and I) Cells were stabled with Annexin V or PI solutions, and cell apoptosis was detected using flow cytometry. (J-L) ELISA of the TNF-α, IL-6 and IL-1β concentrations in HK-2 cells. Data are presented as the mean ± standard deviation from three independent experiments. ***P<0.001 vs. control group; ##P<0.01 and ###P<0.001 vs. H/R + NC inhibitor group; &P<0.05, &&P<0.01 and &&&P<0.001 vs. H/R + miR-328-3p inhibitor + siNC group. PIM1, pim-1 proto-oncogene; NF-2, neurofibromin 2; RT-qPCR, reverse transcription-quantitative PCR; H/R, hypoxia/reperfusion; WT, wild-type; MUT, mutant-type; IL-1β, interleukin-1β; IL-6, interleukin-6; TNF-α, tumor necrosis factor-α.
To examine the effects of PIM1 regulation by miR-328-3p in H/R-exposed cells, siPIM1, miR-328-3p inhibitor and the corresponding control plasmid were generated and transfected into HK-2 cells. The knockdown of miR-328-3p was further confirmed (P<0.001, Fig. S1B). RT-qPCR analysis was also performed to confirm the inhibitory degree of silencing PIM1 (P<0.001, Fig. S1C). Furthermore, the viability, apoptosis and inflammatory potential of H/R-exposed HK-2 cells were assessed. An ~20% decrease in cell viability was observed in the cells in which miR-328-3p was silenced, compared to the H/R-exposed cells (P<0.001, Fig. 2G). However, PIM1 knockdown in the miR-328-3p-silencing cells resulted in a significant increase in cell viability compared to the H/R + miR-328-3p inhibitor group (P<0.05, Fig. 2G). Moreover, it was observed that the re-expression of miR-328-3p induced a significant increase in the apoptosis of H/R-exposed HK-2 cells; it also further increased the levels of pro-inflammatory factors (P<0.001, Fig. 2H-L). In addition, the silencing of PIM1 reversed the cell damage triggered by H/R and miR-328-3p knockdown in HK-2 cells (P<0.001, Fig. 2H-L). These data indicated that PIM1 upregulation by miR-328-3p exerts pro-inflammatory effects in H/R-exposed HK-2 cells.
circITGB1 functions as a sponge of miR-328-3p
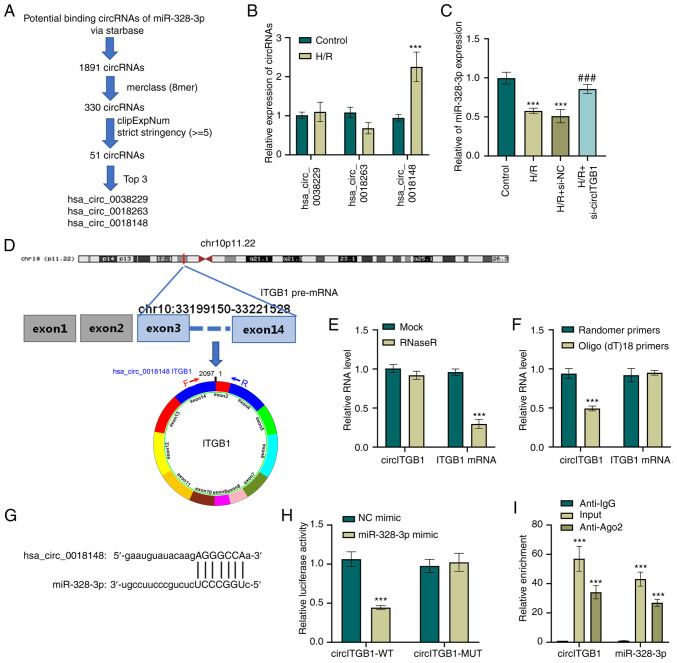
Since circRNAs are aberrantly regulated in human diseases, the present study explored whether the level of miR-328-3p in HK-2 cells could be regulated by specific circRNAs. To this end, circRNAs that have potential binding sites to miRNAs were first examined using StarBase (http://starbase.sysu.edu.cn/). By applying an additional screening parameter (8mer class, clipExpNum strict stringency ≥5), the results were narrowed down to three circRNAs candidates that target miR-328-3p (Fig. 3A). The present study then examined the expression levels of these circRNAs (hsa_circ_0038229, hsa_circ_0018263 and hsa_circ_0018148) in H/R-exposed HK-2 cells using RT-qPCR. The results revealed that H/R resulted in an upregulation of hsa_circ_0018148 (P<0.001, Fig. 3B). To confirm the effect of hsa_circ_0018148 (also known as circITGB1) on the miR-328-3p level in HK-2 cells, siRNA targeting circITGB1 was transfected into the HK-2 cells. RT-qPCR analysis was performed to confirm the inhibitory effect of si-circITGB1 on the circITGB1 expression level (P<0.001, Fig. S1D) and to exclude an off-target effect of the siRNA. miR-328-3p expression was also examined in ITGB1-silenced cells and it was found that the H/R-exposed cells that were transfected with si-circITGB1 expressed a higher level of miR-328-3p when compared to the cells that exposed to H/R (P<0.001, Fig. 3C). hsa_circ_0038229 (2,097 bp sequence in length), is generated from exons 2-14 of the ITGB1 gene located on chr10:33199150-33221528 (Fig. 3D). It was found that circITGB1 was resistant to RNase R digestion, while the linear mRNA of ITGB1 in HK-2 cells was markedly decreased by RNase R treatment (P<0.001, Fig. 3E). Moreover, RT-qPCR was utilized to detect the levels of circITGB1 and ITGB1 mRNA following reverse transcription with random primers and oligo(dt)18 primers (P<0.001, Fig. 3F). Additionally, as shown in Fig. 3G, a target sequence of circITGB1 was found in miR-328-3p, suggesting that circITGB1 directly targeted miR-328-3p. To further confirm this, a fragment of wild-type circITGB1 containing the putative miR-328-3p target sequence was first cloned into a luciferase reporter vector, and it was found that miR-328-3p mimic transfection resulted in a significant decrease in luciferase activity (P<0.001, Fig. 3H). However, the luciferase activity of the circITGB1-MUT + miR-328-3p mimic group had no significant change compared to the circITGB1-MUT + NC mimic group. In addition, RIP assay was performed in H/R-exposed HK-2 cells to precipitate circITGB1 and miR-328-3p with an anti-Ago2 antibody. The data suggested that both circITGB1 and miR-328-3p were evidently pulled down by anti-Aso2 antibody compared with the IgG group (P<0.001, Fig. 3I). Collectively, these results indicated that circITGB1 functioned as a sponge of miR-328-3p in HK-2 cells.
Figure 3.
circITGB1 functions as a sponge of miR-328-3p. (A) Schematic representation of putative miRNA targeting circRNA prediction. (B) HK-2 cells were exposed to with hypoxian (12 h) and reoxygenation for 6 h. Then, the expression levels of hsa_circ_0038229, hsa_circ_0018263 and hsa_circ_0018148 were determined using RT-qPCR. ***P<0.001 vs. control group. (C) Expression levels of miR-328-3p in HK-2 transduced with si-NC or si-circITGB1 and treated with H/R were examined using RT-qPCR analysis. ***P<0.001 vs. control group; ###P<0.001 vs. H/R + si-NC group. (D) Schematic illustration of the formation of circITGB1 via the circularization of exons 3-14 in ITGB1 gene. (E) The expression of circITGB1 and ITGB1 mRNA following treatment with RNase R. ***P<0.001 vs. mock group. (F) Random primers and oligo (dt)18 primers were utilized for reverse transcription and RT-qPCR was performed to examine the expression of circITGB1 and ITGB1 mRNA. ***P<0.001 vs. random primers group. (G) Sequence alignment between miR-328-3p and circITGB1. (H) Luciferase reporter gene assay was carried out with miR-328-3p mimic or NC mimic transfection in cells with co-transfection of circITGB1-WT/MUT. ***P<0.001 vs. NC mimic group. (I) RIP assay performed with to RNA immunoprecipitation assay with anti-Ago2 antibody was performed in H/R-treated HK-2 cells, followed by RT-qPCR to examine the enrichment of circITGB1 and miR-328-3p. Data are presented as the mean ± standard deviation from three independent experiments. ***P<0.001 vs. Anti-IgG group. circITGB1, circular RNA integrin beta 1; RT-qPCR, reverse transcription-quantitative PCR; H/R, hypoxia/reperfusion; WT, wild-type; MUT, mutant-type.
Silencing of circITGB1 reverses the inflammatory response by increasing miR-328-3p expression in H/R-exposed HK-2 cells
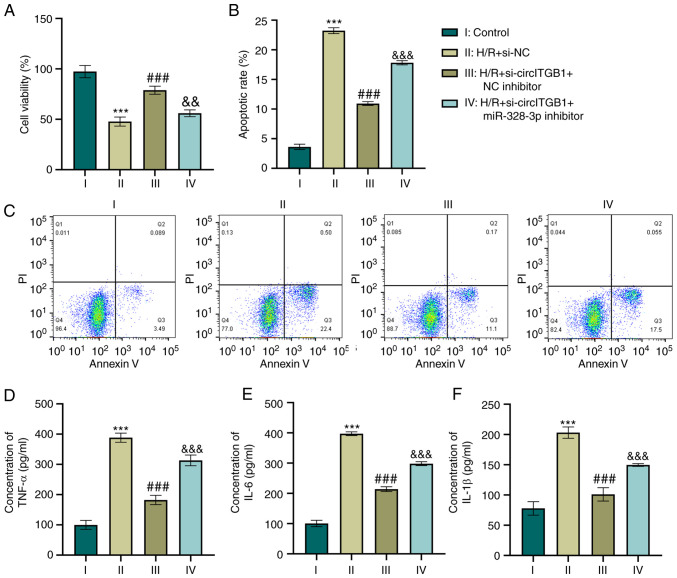
As it was found that circITGB1 functions as a regulator of miR-328-3p expression which modulates cell inflammation, the present study then investigated whether circITGB1 affects the inflammatory context by binding miR-328-3p. The knockdown of circITGB1 alone increased the growth of HK-2 cells compared to the H/R group (P<0.001, Fig. 4A). This was consistent with the reduction of cell apoptosis in the H/R + si-circITGB1 + NC inhibitor group (P<0.001, Fig. 4B and C). Furthermore, the combination of si-circITGB1 and miR-328-3p inhibitor transfection decreased cell viability compared to the co-transfection of si-circITGB1 and NC inhibitor (P<0.01, Fig. 4A). However, the apoptotic percentage was increased in the H/R + si-circITGB1 + miR-328-3p inhibitor group compared with the H/R + si-circITGB1 + NC inhibitor group (P<0.001, Fig. 4B and C). Moreover, the concentrations of pro-inflammatory factors (TNF-α, IL-6 and IL-1β) were decreased following si-circITGB1 transfection compared with the H/R group (P<0.001, Fig. 4D-F). However, the addition of miR-328-3p inhibitor reversed the inhibitory effects of si-circITGB1 on the secretion of inflammatory factors in H/R-exposed cells (P<0.001, Fig. 4D-F).
Figure 4.
Silencing circITGB1 reverses the inflammatory response by increasing miR-328-3p expression in H/R-exposed HK-2 cells. (A) HK-2 cells were co-transfected with miR-328-3p inhibitor/NC inhibitor and si-circITGB1/si-NC and then exposed to H/R. Cell viability was evaluated using CCK-8 assay by detecting the absorbance at 450 nm. (B and C) Cells were stabled with Annexin V or PI solutions, and cell apoptosis was detected using flow cytometry. (D-F) ELISA of the TNF-α, IL-6 and IL-1β concentrations in HK-2 cells. Data are presented as the mean ± standard deviation from three independent experiments. ***P<0.001 vs. control group; ###P<0.001 vs. H/R + si-NC group; &&P<0.01 and &&&P<0.001 vs. H/R + si-circITGB1 + miR-328-3p inhibitor group. circITGB1, circular RNA integrin beta 1; H/R, hypoxia/reperfusion; IL-1β, interleukin-1β; IL-6, interleukin-6; TNF-α, tumor necrosis factor-α.
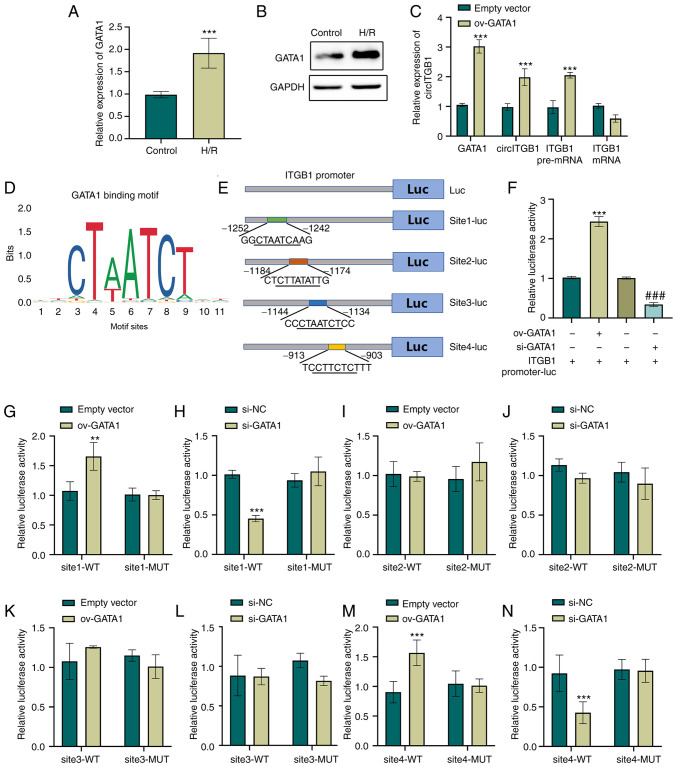
GATA1 is a transcription factor which regulates circITGB1
Accumulating evidence indicates that transcription factors can bind to the promoter of circRNAs and then regulate the effects of circRNAs in human diseases (24,25). To investigate the mechanisms through which circITGB1 is regulated in HK-2 cells, the JASPAR database (http://jaspar.genereg.net) was used to predict potential transcription factors that can bind to the promoter of ITGB1. The predication results indicated four putative GATA1-binding sites upstream of the transcription start site (TSS) (−1,252~−1,242, −1,184~−1,174, −1,144~−1,134 and −913~−903 bp) in the ITGB1 promoter (Fig. 5E). GATA1 is a member of the GATA family of essential transcription factors (26) and modulates gene expression at transcriptional level by binding to specific promoters. Firstly, the present study examined the effect of H/R on GATA1 expression. As shown in Fig. 5A and B, H/R markedly enhanced the GATA1 mRNA and protein expression level in HK-2 cells (P<0.001). To elucidate the effects of GATA1 on the level of circITGB1, GATA1 overexpression vectors were constructed and transfected into HK-2 cells. The successful overexpression of GATA1 in HK-2 cells was confirmed using RT-qPCR (P<0.01, Fig. S1D). In addition, transfecting HK-2 cells with GATA1-specific siRNA leed to a 50% decrease in the GATA1 expression level in the si-GATA1 group compared to that in the si-NC group (P<0.01, Fig. S1G). Subsequently, to verify the transcriptional regulation of GATA1 on ITGB1, the expression of circITGB1, TGB1 mRNA and pre-mRNA was examined in GATA1-overexpressing cells. The results indicated that the exogenous expression of GATA1 increased the expression of circITGB1 (P<0.001, Fig. 5C). Of note, GATA1 overexpression increased the ITGB1 pre-mRNA level (P<0.001), but did not affect the expression of ITGB1 mRNA (Fig. 5C). To confirm the binding of GATA1 to the ITGB1 promoter, the 1.5-kb promoter region and the ITGB1 fragments containing potential binding sites were cloned into a luciferase reporter vector, respectively (Fig. 5D and E). The dual-luciferase reporter assay indicated that GATA1 overexpression increased the ITGB1 promoter activity, whereas GATA1 knockdown decreased ITGB1 promoter activity (P<0.001, Fig. 5F). In addition, the present study further explored which site GATA1 binds to the ITGB1 promoter. The results revealed that the sequences of −1,252~−1,242 and −913~−903 bp upstream of the ITGB1 TSS were essential for the GATA1-mediated transcriptional activation of the ITGB1 gene (P<0.001, Fig. 5G-N). Taken together, these results suggested that ITGB1 is the target gene of transcription factor GATA1.
Figure 5.
GATA1 is a transcription factor which regulates circITGB1. (A and B) HK-2 cells were subjected to hypoxia (12 h) and reoxygenation for 6 h. The expression levels of GATA1 were then determined using RT-qPCR and western blot analysis. ***P<0.001 vs. control group. (C) The expression levels of GATA1, circITGB1, ITGB1 pre-mRNA and ITGB1 mRNA in GATA1-overexpressing HK-2 cells were examined using RT-qPCR. ***P<0.001 vs. empty vector group. (D) JASPAR database prediction of the four putative GATA1 binding motif in ITGB1 promoter. (E) Schematic representation of promoter reporter constructs termed ITGB1 promoter-luc, site1-luc, site2-luc, site3-luc and site4-luc. (F) Relative luciferase activity was detected in HK-2 cells co-transfected with luciferase plasmids containing ITGB1 promoter sequences and ov-GATA1/si-GATA1. ***P<0.001 vs. ITGB1 promoter-luc group; ###P<0.001 vs. ITGB1 promoter-luc group (G-N) HK-2 cells were co-transfected with wild-type or mutant ITGB1 promoter luciferase constructs containing GATA1 binding sites and GATA1 overexpressing vectors or si-GATA1. Luciferase reporter assay was performed to confirm the binding sites. Data are presented as the mean ± standard deviation from three independent experiments. **P<0.01 and ***P<0.001 vs. empty vector group or si-NC. GATA1, GATA binding protein 1; circITGB1, circular RNA integrin beta 1; RT-qPCR, reverse transcription-quantitative PCR; WT, wild-type; MUT, mutant-type.
Discussion
AKI is a fatal renal disease due to its late detection and reperfusion-induced irreversible damage during kidney transplantation surgery (27,28). Renal tubules are the first to be damaged under hypoxic or toxic conditions and induce interstitial inflammation and immune responses (29,30). Primary renal proximal tubule epithelial cells (RPTECs) are valuable cell models for the study of IRI in vitro (31). However, it is difficult to ensure the highest viability and plating efficiency of RPTECs during cell culture. Human tubular epithelial cells (HK-2) are an immortalized cell line derived from proximal convoluted tubules of normal kidneys (32). It has been reported that in vitro IRI HK-2 cell models were induced by hypoxia in anaerobic chambers (33). The present study found that the overexpression of miR-328-3p reduced cell apoptosis and the inflammatory response of H/R-exposed HK-2 cells by targeting the PIM1 3′UTR. This result suggested that there was a lower expression of miR-328-3p or a higher level of PIM1 following H/R that could account for renal injury. Furthermore, it was found that cell inflammatory injury was mediated by the overexpression of circITGB1 and the transcription factor GATA1 may bind to the ITGB1 promoter, resulting in the expression of circITGB1.
The late detection of AKI patients is dependent not only on the urine output criteria or serum creatinine criteria (28), but also on the specific exosome microenvironment (34). With respect to this, several exosome miRNAs exert considerable functions in the inflammasome activation and cascade. Hence, the identification of the exosome miRNAs and the confirmation of their role in IRI may offer specific therapeutic intervention strategies for AKI. The present study found that exosome rno-miR-328a-3p was downregulated in IRI datasets. Moreover, rno-miR-328a-3p is homologous with hsa-miR-328-3p. Thus, HK-2 cells were used to induce IRI and explore the role of miR-328-3p in the human cell inflammatory response. miR-328 has been shown to regulate the MEK-ERK signaling pathway, resulting in the inhibition of myocardial IRI and apoptosis (35). The findings of the present study suggested that the overexpression of miR-328-3p resulted in a decrease in PIM1 expression and, therefore, played an anti-inflammatory role in H/R-exposed HK-2 cells.
PIM1 belongs to the Ser/Thr protein kinase family and functions as a proto-oncogene in human malignancies (36). PIM1 plays a role in the regulation of cell proliferation and survival in breast cancer (37), prostate cancer (38) and colorectal cancer (39). A role in hypoxia environment has been described for PIM1, since the apoptosis of rat cardiomyocytes was inhibited by PIM1 overexpression to enhance autophagy in spite of oxidative stress (40). Accordingly, the present found that PIM1 expression was upregulated in H/R-exposed HK-2 cells, and its knockdown reversed the pro-apoptotic and pro-inflammatory effects of miR-328-3p inhibitor.
Non-coding RNAs, particularly circRNAs (41), have been linked to the regulation of miRNAs. It is considered that the circRNA-miRNA axis is effective in the regulation of human diseases, and is able to initiate or inhibit mRNA expression and modulate biological functions (42). Indeed, a previous study by the authors indicated that circYAP1 sponged miR-21-5p and prevented HK-2 cells from IRI (43). As miR-328-3p expression was downregulated in H/R-exposed cells, it was hypothesized that circRNAs which are overexpressed following H/R may be candidates for sponging miR-328-3p in AKI. CircInteractome can generate 21-nt siRNAs targeting junctional sequences spanning 5-16 nt on either side of the junction. The 21-nt siRNAs (GUGGAGAAUCCAGAUGAAAAU) are specific to the mature sequence of hsa_circ_0018148. This siRNA specifically knocks down the hsa_circ_0018148 without affecting the linear counterpart mRNA. BLAST analysis directed from CircInteractome revealed that the siRNA sequence targets UBR4 with 16 bp complementation (siRNA nucleotides 2-17). To exclude an off-target effect of siRNA on hsa_circ_0018148, the expression of the UBR4 gene was detected, which was shown to exhibit significant alignment with siRNA by BLAST analysis. RT-qPCR analysis revealed that the siRNA did not significantly decrease the UBR4 expression level in HK-2 cells (Fig. S1E). A possible explanation for this may be that the potential target UBR4 mRNA could form several complex RNA secondary structures around the siRNA matching sites. The formed RNA secondary structures will influence the complementary base pairing between siRNA antisense strand and targeting sequence, thus influencing the RNAi efficiency (44). In the present study, it was confirmed that the knockdown of circITGB1 not only increased the expression level of miR-328-3p, but also led to decreased cell inflammatory properties following exposure to H/R. Nevertheless, the upstream mechanism of circITGB1 overexpression in AKI remains to be elucidated.
Previous studies have established that transcription factors may be related to gene or circRNA regulation (45,46). For example, it has been shown that transcription factor YY1 can bind to the promoter of circ-ZNF609 and regulate the progression in cholangiocarcinoma (47). The promoter of circHipk2 has also been found to be regulated by transcription factor Sp1 and circHipk2 is regarded as an important candidate regulating myogenesis (48). The transcriptional activity of circITGB1 derives from the main promoter region upstream of ITGB1 coding sequence, which includes several transcription factor binding sites (46). In the present study, to determine the upstream regulatory mechanism of circITGB1 under H/R conditions, bioinformatics analysis indicated that the transcription factor GATA1 could bind to four sites region on the ITGB1 promoter sequence.
GATA1 contains two zinc finger domain, an N-terminal and a C-terminal transactivation domain (49). Emerging evidence indicates GATA1 is activated in several tumors, such as prostate, colorectal and renal cancer (50). However, the functions of GATA1 in AKI and IRI have not been reported to date, at least to the best of our knowledge. GATA1 encodes a key hematopoietic transcription factor essential for the specification of erythroid cells from early hematopoietic stem and progenitor cells. A lack of GATA1 activity can be a major pathogenic mechanism that contributes to anemia in vivo. Under hypoxic conditions, the kidneys will produce erythropoietin (EPO), which is carried through the blood to the bone marrow and spleen to enhance proliferation and survival of erythroid progenitors to ablate the hypoxic conditions, and reduce EPO production back to normal levels (51). The transcription factor GATA1 appears to be specifically upregulated during the course of erythroid differentiation (52,53). In addition, GATA1 exerts a regulatory function in promoting pro-inflammatory cytokines levels by mediating nitric oxide synthase 2 transcription (54). Inflammation is a common pathophysiological manifestation of clinical AKI or IRI in vivo (55). In the present study, the results indicated that H/R increased the expression level of GATA1 in HK-2 cells. In further mechanistic analyses, GATA1 was found to regulate ITGB1 transcription by binding to specific consensus binding sites (CTAATCT) within the ITGB1 promoter. Specifically, putative binding sites for GATA1 are located at nucleotides −1.252/−1.242 and −913/−903 upstream of the transcription start site within ITGB1 promoter. To the best of our knowledge, the present study was the first to demonstrate that GATA1 contributed to the aberrant increase in the expression of circITGB1.
It is worth noting that there were some limitations to the present study. Firstly, in vivo IRI samples need to be analyzed to confirm the expression levels of circITGB1/miR-328-3p. Secondly, the distribution of circITGB1 expression in the cytoplasm or nucleus in H/R-exposed HK-2 cells remains to be investigated in order to understand the transcriptional regulation. Thirdly, the pathway downstream PIM1 needs to be further explored to clarify the mechanisms involved in IRI.
In conclusion, the present study demonstrated a novel pathway which places circITGB1/miR-328-3p/PIM1 under the transcriptional control of GATA1 in H/R-treated HK-2 cells. The findings presented herein confirmed the pro-inflammatory role of circITGB1 in IRI, suggesting potential targets with the miR-328-3p/PIM1 axis during the progression of AKI. Further studies are required however, to evaluate the importance of this regulatory axis in vivo.
Supplementary Data
Acknowledgements
Not applicable.
Abbreviations
- AKI
acute kidney injury;
- IRI
ischemia/reperfusion injury;
- H/R
hypoxia/reperfusion;
- circRNA
circularRNA;
- miRNA/miR
microRNA;
- 3′UTR
3′untranslated region;
- ITGB1
integrin beta 1;
- PIM1
pim-1 proto-oncogene;
- GATA1
GATA binding protein 1;
- HK-2 cell
human kidney-2 cell;
- K-SFM
keratinocyte serum free medium;
- siRNA
small interfering RNA;
- RT-qPCR
reverse transcription-quantitative PCR;
- CCK-8
Cell Counting Kit-8;
- IL-1β
interleukin-1β
- IL-6
interleukin-6;
- TNF-α
tumor necrosis factor-α
- HRP
horseradish peroxidase;
- RIP
RNA immunoprecipitation;
- SD
standard deviation;
- ANOVA
analysis of variance;
- FC
fold change;
- TSS
transcription start site;
- EPO
erythropoietin
Funding Statement
The present study was supported by the Natural Science Foundation of Shandong Province of China (grant no. ZR2020QH237).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus (GEO) repository (GSE124669; https://www.ncbi.nlm.nih.gov/gds).
Authors' contributions
TH conceptualized the study, provided the study resources, supervised the study, acquired funding and confirm the authenticity of all the raw data. YG and TH were involved in performing experiments, data curation, as well as in preparing the table and figures, methodology, in the writing of the original draft, project administration and in the writing, reviewing and editing of the manuscript. YG, WJX and CG performed the bioinformatics analysis and were involved in the formal analysis. YG was involved in the investigative aspects of the study. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Pesavento TE. Kidney transplantation in the context of renal replacement therapy. Clin J Am Soc Nephrol. 2009;4:2035–2039. doi: 10.2215/CJN.05500809. [DOI] [PubMed] [Google Scholar]
- 2.Kadir S, Watson A, Burrow C. Percutaneous transcatheter recanalization in the management of acute renal failure due to sudden occlusion of the renal artery to a solitary kidney. Am J Nephrol. 1987;7:445–449. doi: 10.1159/000167517. [DOI] [PubMed] [Google Scholar]
- 3.Bahl D, Haddad Z, Datoo A, Qazi YA. Delayed graft function in kidney transplantation. Curr Opin Organ Transplant. 2019;24:82–86. doi: 10.1097/MOT.0000000000000604. [DOI] [PubMed] [Google Scholar]
- 4.Tammaro A, Kers J, Scantlebery AML, Florquin S. Metabolic flexibility and innate immunity in renal ischemia reperfusion injury: The fine balance between adaptive repair and tissue degeneration. Front Immunol. 2020;11:1346. doi: 10.3389/fimmu.2020.01346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang L, Xing G, Wang L, Wu Y, Li S, Xu G, He Q, Chen J, Chen M, Liu X, et al. Acute kidney injury in China: A cross-sectional survey. Lancet. 2015;386:1465–1471. doi: 10.1016/S0140-6736(15)00344-X. [DOI] [PubMed] [Google Scholar]
- 6.Catalanotto C, Cogoni C, Zardo G. MicroRNA in control of gene expression: An overview of nuclear functions. Int J Mol Sci. 2016;17:1712. doi: 10.3390/ijms17101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hou P, Li H, Yong H, Chen F, Chu S, Zheng J, Bai J. PinX1 represses renal cancer angiogenesis via the mir-125a-3p/VEGF signaling pathway. Angiogenesis. 2019;22:507–519. doi: 10.1007/s10456-019-09675-z. [DOI] [PubMed] [Google Scholar]
- 8.Fan Y, Chen H, Huang Z, Zheng H, Zhou J. Emerging role of miRNAs in renal fibrosis. RNA Biol. 2020;17:1–12. doi: 10.1080/15476286.2019.1667215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramanathan K, Padmanabhan G. miRNAs as potential biomarker of kidney diseases: A review. Cell Biochem Funct. 2020;38:990–1005. doi: 10.1002/cbf.3555. [DOI] [PubMed] [Google Scholar]
- 10.Song N, Zhang T, Xu X, Lu Z, Yu X, Fang Y, Hu J, Jia P, Teng J, Ding X. miR-21 Protects against ischemia/reperfusion-induced acute kidney injury by preventing epithelial cell apoptosis and inhibiting dendritic cell maturation. Front Physiol. 2018;9:790. doi: 10.3389/fphys.2018.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He W, Zhuang J, Zhao ZG, Luo H, Zhang J. miR-328 prevents renal fibrogenesis by directly targeting TGF-β2. Bratisl Lek Listy. 2018;119:434–440. doi: 10.4149/BLL_2018_079. [DOI] [PubMed] [Google Scholar]
- 12.Shi Y, Jia X, Xu J. The new function of circRNA: Translation. Clin Transl Oncol. 2020;22:2162–2169. doi: 10.1007/s12094-020-02371-1. [DOI] [PubMed] [Google Scholar]
- 13.Hsiao KY, Sun HS, Tsai SJ. Circular RNA-new member of noncoding RNA with novel functions. Exp Biol Med (Maywood) 2017;242:1136–1141. doi: 10.1177/1535370217708978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wei W, Yao Y, Bi H, Xu W, Gao Y. Circular RNA circ_0068,888 protects against lipopolysaccharide-induced HK-2 cell injury via sponging microRNA-21-5p. Biochem Biophys Res Commun. 2021;540:1–7. doi: 10.1016/j.bbrc.2020.12.018. [DOI] [PubMed] [Google Scholar]
- 15.Kölling M, Seeger H, Haddad G, Kistler A, Nowak A, Faulhaber-Walter R, Kielstein J, Haller H, Fliser D, Mueller T, et al. The circular RNA ciRs-126 predicts survival in critically Ill patients with acute kidney injury. Kidney Int Rep. 2018;3:1144–1152. doi: 10.1016/j.ekir.2018.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 17.Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sonoda H, Lee BR, Park KH, Nihalani D, Yoon JH, Ikeda M, Kwon SH. miRNA profiling of urinary exosomes to assess the progression of acute kidney injury. Sci Rep. 2019;9:4692. doi: 10.1038/s41598-019-40747-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romualdi C, Bortoluzzi S, D'Alessi F, Danieli GA. IDEG6: A web tool for detection of differentially expressed genes in multiple tag sampling experiments. Physiol Genomics. 2003;12:159–162. doi: 10.1152/physiolgenomics.00096.2002. [DOI] [PubMed] [Google Scholar]
- 20.Guo C, Dong G, Liang X, Dong Z. Epigenetic regulation in AKI and kidney repair: Mechanisms and therapeutic implications. Nat Rev Nephrol. 2019;15:220–239. doi: 10.1038/s41581-018-0103-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fan PC, Chen CC, Chen YC, Chang YS, Chu PH. MicroRNAs in acute kidney injury. Hum Genomics. 2016;10:29. doi: 10.1186/s40246-016-0085-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding C, Ding X, Zheng J, Wang B, Li Y, Xiang H, Dou M, Qiao Y, Tian P, Xue W. miR-182-5p and miR-378a-3p regulate ferroptosis in I/R-induced renal injury. Cell Death Dis. 2020;11:929. doi: 10.1038/s41419-020-03135-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pu M, Chen J, Tao Z, Miao L, Qi X, Wang Y, Ren J. Regulatory network of miRNA on its target: Coordination between transcriptional and post-transcriptional regulation of gene expression. Cell Mol Life Sci. 2019;76:441–451. doi: 10.1007/s00018-018-2940-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng L, Liu S, Liu F, Sang M, Ju Y, Fan X, Gu L, Li Z, Geng C, Sang M. ZEB1-mediated transcriptional upregulation of circWWC3 promotes breast cancer progression through activating ras signaling pathway. Mol Ther Nucleic Acids. 2020;22:124–137. doi: 10.1016/j.omtn.2020.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meng J, Chen S, Han JX, Qian B, Wang XR, Zhong WL, Qin Y, Zhang H, Gao WF, Lei YY, et al. Twist1 regulates vimentin through Cul2 circular RNA to promote EMT in hepatocellular carcinoma. Cancer Res. 2018;78:4150–4162. doi: 10.1158/0008-5472.CAN-17-3009. [DOI] [PubMed] [Google Scholar]
- 26.Tremblay M, Sanchez-Ferras O, Bouchard M. GATA transcription factors in development and disease. Development. 2018;145:dev164384. doi: 10.1242/dev.164384. [DOI] [PubMed] [Google Scholar]
- 27.Oh DJ. A long journey for acute kidney injury biomarkers. Ren Fail. 2020;42:154–165. doi: 10.1080/0886022X.2020.1721300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Koeze J, Keus F, Dieperink W, van der Horst IC, Zijlstra JG, van Meurs M. Incidence, timing and outcome of AKI in critically ill patients varies with the definition used and the addition of urine output criteria. BMC Nephrol. 2017;18:70. doi: 10.1186/s12882-017-0487-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu BC, Tang TT, Lv LL, Lan HY. Renal tubule injury: A driving force toward chronic kidney disease. Kidney Int. 2018;93:568–579. doi: 10.1016/j.kint.2017.09.033. [DOI] [PubMed] [Google Scholar]
- 30.Zuk A, Bonventre JV. Acute kidney injury. Annu Rev Med. 2016;67:293–307. doi: 10.1146/annurev-med-050214-013407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vormann MK, Tool LM, Ohbuchi M, Gijzen L, van Vught R, Hankemeier T, Kiyonaga F, Kawabe T, Goto T, Fujimori A, et al. Modelling and prevention of acute kidney injury through ischemia and reperfusion in a combined human renal proximal tubule/blood vessel-on-a-chip. Kidney360. 2021;3:217–231. doi: 10.34067/KID.0003622021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ryan MJ, Johnson G, Kirk J, Fuerstenberg SM, Zager RA, Torok-Storb B. HK-2: An immortalized proximal tubule epithelial cell line from normal adult human kidney. Kidney Int. 1994;45:48–57. doi: 10.1038/ki.1994.6. [DOI] [PubMed] [Google Scholar]
- 33.Zhang G, Yang Y, Huang Y, Zhang L, Ling Z, Zhu Y, Wang F, Zou X, Chen M. Hypoxia-induced extracellular vesicles mediate protection of remote ischemic preconditioning for renal ischemia-reperfusion injury. Biomed Pharmacother. 2017;90:473–478. doi: 10.1016/j.biopha.2017.03.096. [DOI] [PubMed] [Google Scholar]
- 34.Thongboonkerd V. Roles for exosome in various kidney diseases and disorders. Front Pharmacol. 2020;10:1655. doi: 10.3389/fphar.2019.01655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ye HK, Zhang HH, Tan ZM. miR-328 inhibits cell apoptosis and improves cardiac function in rats with myocardial ischemia-reperfusion injury through MEK-ERK signaling pathway. Eur Rev Med Pharmacol Sci. 2020;24:3315–3321. doi: 10.26355/eurrev_202003_20699. [DOI] [PubMed] [Google Scholar]
- 36.Merkel AL, Meggers E, Ocker M. PIM1 kinase as a target for cancer therapy. Expert Opin Investig Drugs. 2012;21:425–436. doi: 10.1517/13543784.2012.668527. [DOI] [PubMed] [Google Scholar]
- 37.Brasó-Maristany F, Filosto S, Catchpole S, Marlow R, Quist J, Francesch-Domenech E, Plumb DA, Zakka L, Gazinska P, Liccardi G, et al. PIM1 kinase regulates cell death, tumor growth and chemotherapy response in triple-negative breast cancer. Nat Med. 2016;22:1303–1313. doi: 10.1038/nm.4198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang C, Qie Y, Yang T, Wang L, Du E, Liu Y, Xu Y, Qiao B, Zhang Z. Kinase PIM1 promotes prostate cancer cell growth via c-Myc-RPS7-driven ribosomal stress. Carcinogenesis. 2019;40:52–60. doi: 10.1093/carcin/bgy126. [DOI] [PubMed] [Google Scholar]
- 39.Li Q, Chen L, Luo C, Yan Chen, Ge J, Zhu Z, Wang K, Yu X, Lei J, Liu T, et al. TAB3 upregulates PIM1 expression by directly activating the TAK1-STAT3 complex to promote colorectal cancer growth. Exp Cell Res. 2020;391:111975. doi: 10.1016/j.yexcr.2020.111975. [DOI] [PubMed] [Google Scholar]
- 40.Zhu HH, Wang XT, Sun YH, He WK, Liang JB, Mo BH, Li L. Pim1 overexpression prevents apoptosis in cardiomyocytes after exposure to hypoxia and oxidative stress via upregulating cell autophagy. Cell Physiol Biochem. 2018;49:2138–2150. doi: 10.1159/000493817. [DOI] [PubMed] [Google Scholar]
- 41.Beermann J, Piccoli MT, Viereck J, Thum T. Non-coding RNAs in development and disease: Background, mechanisms, and therapeutic approaches. Physiol Rev. 2016;96:1297–1325. doi: 10.1152/physrev.00041.2015. [DOI] [PubMed] [Google Scholar]
- 42.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–691. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 43.Huang T, Cao Y, Wang H, Wang Q, Ji J, Sun X, Dong Z. Circular RNA YAP1 acts as the sponge of microRNA-21-5p to secure HK-2 cells from ischaemia/reperfusion-induced injury. J Cell Mol Med. 2020;24:4707–4715. doi: 10.1111/jcmm.15142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Overhoff M, Alken M, Far RK, Lemaitre M, Lebleu B, Sczakiel G, Robbins I. Local RNA target structure influences siRNA efficacy: A systematic global analysis. J Mol Biol. 2005;348:871–881. doi: 10.1016/j.jmb.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 45.Hu M, Yu B, Zhang B, Wang B, Qian D, Li H, Ma J, Liu DX. Human cytomegalovirus infection activates glioma activating transcription factor 5 via microRNA in a stress-induced manner. ACS Chem Neurosci. 2021;12:3947–3956. doi: 10.1021/acschemneuro.1c00576. [DOI] [PubMed] [Google Scholar]
- 46.Hu M, Li H, Xie H, Fan M, Wang J, Zhang N, Ma J, Che S. ELF1 transcription factor enhances the progression of glioma via ATF5 promoter. ACS Chem Neurosci. 2021;12:1252–1261. doi: 10.1021/acschemneuro.1c00070. [DOI] [PubMed] [Google Scholar]
- 47.Guan C, Liu L, Zhao Y, Zhang X, Liu G, Wang H, Gao X, Zhong X, Jiang X. YY1 and eIF4A3 are mediators of the cell proliferation, migration and invasion in cholangiocarcinoma promoted by circ-ZNF609 by targeting miR-432-5p to regulate LRRC1. Aging (Albaany NY) 2021;13:25195–25212. doi: 10.18632/aging.203735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yan J, Yang Y, Fan X, Tang Y, Tang Z. Sp1-mediated circRNA circHipk2 regulates myogenesis by targeting ribosomal protein Rpl7. Genes (Basel) 2021;12:696. doi: 10.3390/genes12050696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hasegawa A, Shimizu R. GATA1 activity governed by configurations of cis-acting elements. Front Oncol. 2017;6:269. doi: 10.3389/fonc.2016.00269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peters I, Dubrowinskaja N, Tezval H, Kramer MW, von Klot CA, Hennenlotter J, Stenzl A, Scherer R, Kuczyk MA, Serth J. Decreased mRNA expression of GATA1 and GATA2 is associated with tumor aggressiveness and poor outcome in clear cell renal cell carcinoma. Target Oncol. 2015;10:267–275. doi: 10.1007/s11523-014-0335-8. [DOI] [PubMed] [Google Scholar]
- 51.Haase VH. Regulation of erythropoiesis by hypoxia-inducible factors. Blood Rev. 2013;27:41–53. doi: 10.1016/j.blre.2012.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liang R, Ghaffari S. Advances in understanding the mechanisms of erythropoiesis in homeostasis and disease. Br J Haematol. 2016;174:661–673. doi: 10.1111/bjh.14194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ludwig LS, Gazda HT, Eng JC, Eichhorn SW, Thiru P, Ghazvinian R, George TI, Gotlib JR, Beggs AH, Sieff CA, et al. Altered translation of GATA1 in Diamond-Blackfan anemia. Nat Med. 2014;20:748–753. doi: 10.1038/nm.3557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Liu H, Wei SP, Zhi LQ, Liu LP, Cao TP, Wang SZ, Chen QP, Liu D. Synovial GATA1 mediates rheumatoid arthritis progression via transcriptional activation of NOS2 signaling. Microbiol Immunol. 2018;62:594–606. doi: 10.1111/1348-0421.12637. [DOI] [PubMed] [Google Scholar]
- 55.Sato Y, Yanagita M. Immune cells and inflammation in AKI to CKD progression. Am J Physiol Renal Physiol. 2018;315:F1501–F1512. doi: 10.1152/ajprenal.00195.2018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the Gene Expression Omnibus (GEO) repository (GSE124669; https://www.ncbi.nlm.nih.gov/gds).