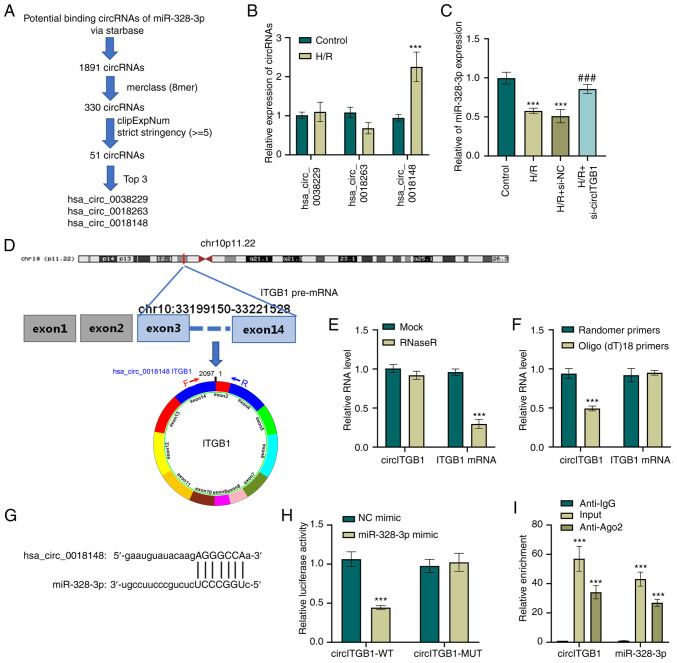
Figure 3.
circITGB1 functions as a sponge of miR-328-3p. (A) Schematic representation of putative miRNA targeting circRNA prediction. (B) HK-2 cells were exposed to with hypoxian (12 h) and reoxygenation for 6 h. Then, the expression levels of hsa_circ_0038229, hsa_circ_0018263 and hsa_circ_0018148 were determined using RT-qPCR. ***P<0.001 vs. control group. (C) Expression levels of miR-328-3p in HK-2 transduced with si-NC or si-circITGB1 and treated with H/R were examined using RT-qPCR analysis. ***P<0.001 vs. control group; ###P<0.001 vs. H/R + si-NC group. (D) Schematic illustration of the formation of circITGB1 via the circularization of exons 3-14 in ITGB1 gene. (E) The expression of circITGB1 and ITGB1 mRNA following treatment with RNase R. ***P<0.001 vs. mock group. (F) Random primers and oligo (dt)18 primers were utilized for reverse transcription and RT-qPCR was performed to examine the expression of circITGB1 and ITGB1 mRNA. ***P<0.001 vs. random primers group. (G) Sequence alignment between miR-328-3p and circITGB1. (H) Luciferase reporter gene assay was carried out with miR-328-3p mimic or NC mimic transfection in cells with co-transfection of circITGB1-WT/MUT. ***P<0.001 vs. NC mimic group. (I) RIP assay performed with to RNA immunoprecipitation assay with anti-Ago2 antibody was performed in H/R-treated HK-2 cells, followed by RT-qPCR to examine the enrichment of circITGB1 and miR-328-3p. Data are presented as the mean ± standard deviation from three independent experiments. ***P<0.001 vs. Anti-IgG group. circITGB1, circular RNA integrin beta 1; RT-qPCR, reverse transcription-quantitative PCR; H/R, hypoxia/reperfusion; WT, wild-type; MUT, mutant-type.