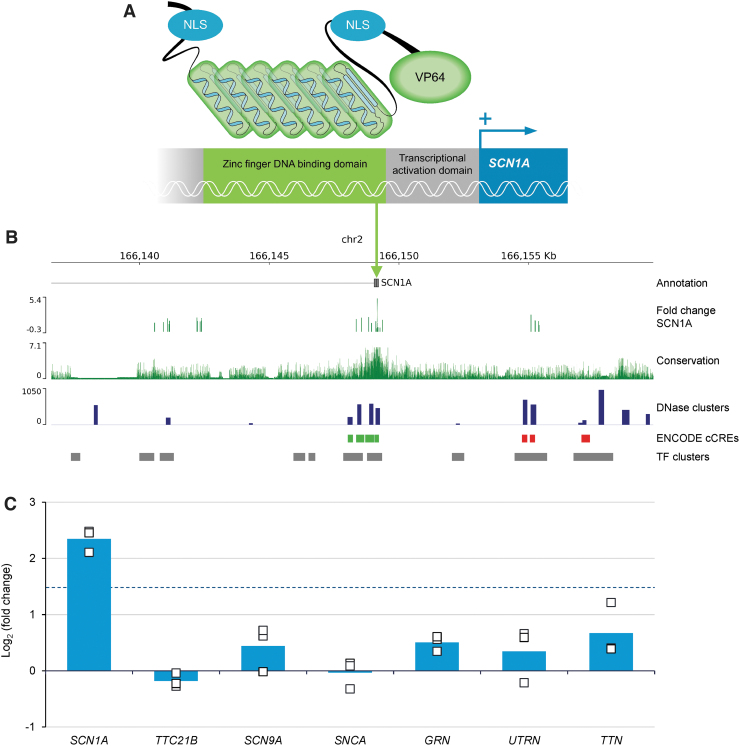
Figure 1.
Structure of eTFSCN1A and its targeted activity at a unique and highly conserved 18-bp noncoding DNA sequence for gene-specific upregulation of SCN1A. (A) Schematic of eTFSCN1A structure. eTFSCN1A is composed of a polydactyl zinc finger DNA-binding domain, a short linker sequence, and a C-terminal VP64 transcriptional activation domain.26,28 Each of the six zinc finger domains interacts with a specific triplet nucleotide, conferring specific interaction with an 18-bp target sequence. NLS domains derived from simian vacuolating virus (SV40) and nucleoplasmin are included at the N-terminal and linker domains. (B) Genomic features of eTFSCN1A target site. Human genome track at the SCN1A locus. Tracks indicate human SCN1A gene annotation, chromosome 2 coordinates (hg38 genome assembly). FC SCN1A track shows target vector candidate screening data in HEK293cells. Mean Log2-FCs in SCN1A expression following transient transfection of eTFSCN1A candidates targeted against different genomic targets are plotted by target coordinates. The eTFSCN1A 18-bp target sequence, which induced the strongest upregulation of SCN1A within the screening window, is indicated by the green arrow. Sequence conservation track is indicated by 100-species base-wise conservation score (PhyloP).29 Regulatory element feature tracks are indicated (UCSC genome browser, ENCODE consortia31,32). DNase cluster track indicates DNase I hypersensitivity cluster score; ENCODE cCREs track marks candidate cis-regulatory element regions (green: PLS, red: dELS); TF clusters track marks transcription factor binding peak clusters. (C) eTFSCN1A upregulates SCN1A in a gene-specific manner in vitro. Change in endogenous gene expression in HEK293 cells is reported in the eTFSCN1A transfected condition relative to control condition for each replicate. Bars represent mean Log2-FC for n = 3 biological replicates (n = 2 for PAPBC1), dots indicate individual replicate measurements. Dotted line indicates Log2FC = 1.5. bp, base-pair; cCREs, candidate cis-regulatory elements; dELS, distal enhancer-like signature; FC, fold change; NLS, nuclear localization signal; PLS, promoter-like signature; TF, transcription factor.