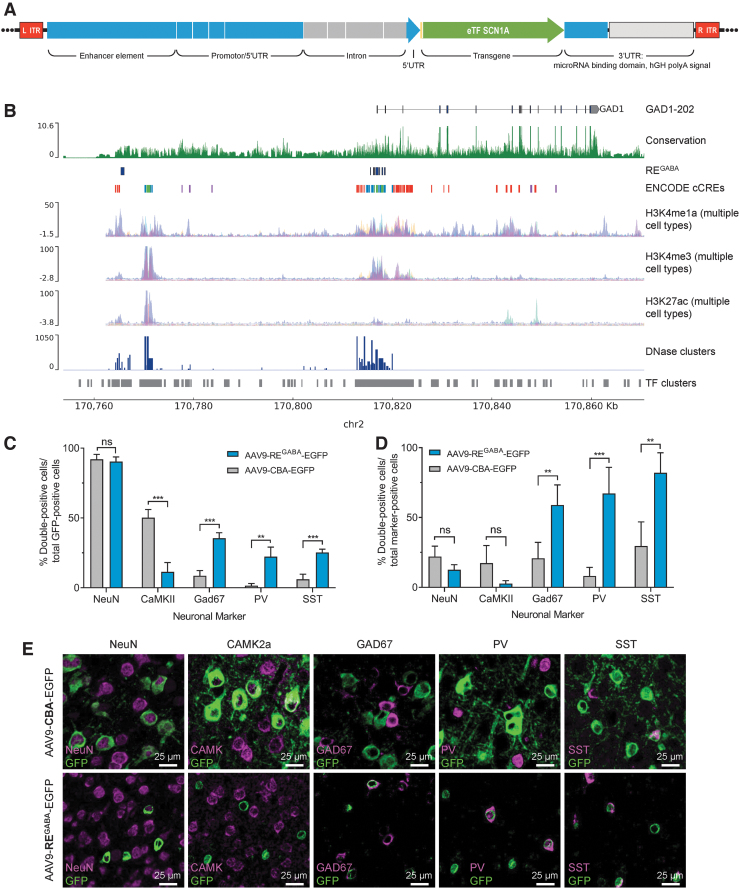
Figure 2.
The REGABA regulatory element in AAV9-REGABA-eTFSCN1A selectively targets GABAergic interneurons in vivo. (A) REGABA construct design. REGABA comprised upstream and 5′UTR sequence components, and a downstream 3′UTR sequence. The upstream regulatory sequence is derived from human genomic sequence elements: an enhancer element located ∼50 kb upstream of the GAD1 gene locus, and proximal promoter and 5′UTR element sequence derived from the human GAD1 gene locus. The promoter is composed of genomic sequence segments in the proximal promoter and 5′UTR of the hGAD1 gene. To enhance gene expression40 and to capture additional regulatory features derived from intronic elements, this promoter also incorporates a truncated intron derived from the first intron of the GAD1 gene, which includes sequences flanking the splice donor and acceptor sites, and internal intronic sequences that overlap high-conservation predicted regulatory sequences. The 3′UTR element includes a collection of eight cognate target motifs derived from excitatory neuron-enriched miRNAs miR128 and mir221,36,39 and a hGH-pA. (B) Genomic features of REGABA source sequence. Human genome track at the human GAD1 locus. Tracks indicate GAD1 gene annotation, chromosome 2 coordinates (hg38 genome assembly). Sequence conservation (PhyloP29,30), and ENCODE regulatory element feature tracks are indicated, as well as additional composite epigenetic marker tracks for histone markers H3K4me1, H3K4me3, and H3K27ac, which can signal active enhancer and promoter regions (UCSC genome browser, ENCODE consortia). (C–E) Expression of EGFP in vivo 27 days post-ICV injection of AAV9-EGFP vectors driven by CBA or REGABA promoters in PND1 mice. Animals (n = 4/group) were administered 2.0E10 vg per animal of AAV-CBA-EGFP or AAV-REGABA-EGFP on PND1 by bilateral ICV injection. On PND28, brain sections were analyzed by IHC for the presence of GFP and neuron-specific markers. Quantitation of colocalization between neuron-specific markersa and EGFP: (C) among GFP-positive cells and (D) compared to the total number of neuron-marker positive cells; **p < 0.01; ***p < 0.001; ns, p ≥ 0.05 (unpaired t-tests, two-stage step-up FDR = 1%; data shown are means and error bars represent SD). (E) Representative images demonstrating colocalization of EGFP driven by each vector with each of the neuronal markers evaluated. aNeuron-specific markers: NeuN (neurons); CAMK2a (glutamatergic neurons); GAD67 (GABAergic interneurons); PV (parvalbumin-positive interneurons); SST (somatostatin-positive interneurons). AAV, adeno-associated virus; CBA, chicken β-actin promoter; chr2, chromosome 2; EGFP, enhanced green fluorescent protein; eTF, engineered transcription factor; hGH-pA, human growth hormone-derived poly-adenylation signal; IHC, immunohistochemistry; ICV, intracerebroventricular; ITR, inverted terminal repeat; ns, not significant; PND, postnatal day; REGABA, GABAergic cell-selective regulatory element; SD, standard deviation; UTR, untranslated region; vg, vector genomes.