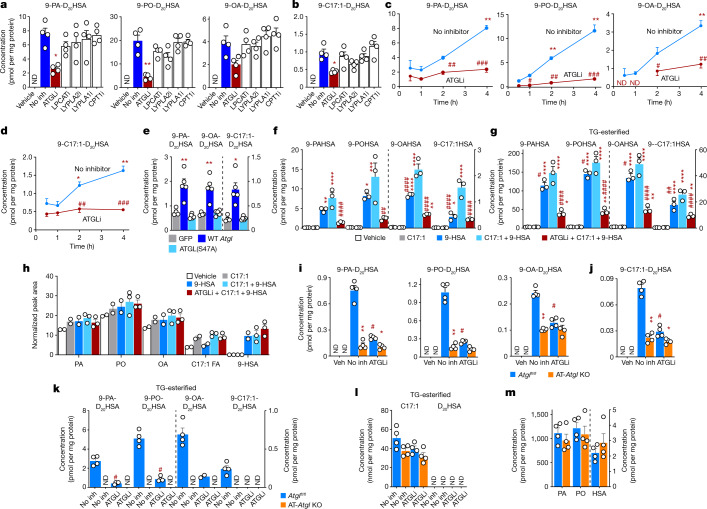
Fig. 2. ATGL regulates biosynthesis of FAHFA and TG-esterified FAHFA in intact cells.
Cells were incubated with both C17:1 FA and D20-9-HSA (20 µM (a–d, i–l) or 25 µM (e–h) each) or 0.05% DMSO (vehicle) for 4 h. a, b, Biosynthesis of 9-PA-D20HSA, 9-PO-D20HSA and 9-OA-D20HSA (a) and 9-C17:1-D20HSA (b) was measured in WT SVF adipocytes pre-incubated with the indicated enzyme inhibitors or 0.1% DMSO (No inh). LPCATi, lysophosphatidylcholine acyltransferase inhibitor; LYPLA2i, lysophospholipase 2 inhibitor; LYPLA1i, lysophospholipase 1 inhibitor; CPT1i, carnitine palmitoyltransferase 1 inhibitor. n = 4 wells; mean ± s.e.m. *P < 0.001, **P < 0.0001 versus No inh group (one-way ANOVA). c, d, Biosynthesis of 9-PA-D20HSA, 9-PO-D20HSA and 9-OA-D20HSA (c) and 9-C17:1-D20HSA (d) in WT SVF adipocytes co-incubated with atglistatin (ATGL inhibitor (ATGLi), 10 µM) or 0.1% DMSO (No inh) for 0.5–4 h. n = 4; mean ± s.e.m. *P < 0.05, **P < 0.0001 versus 0.5 h, same treatment, #P < 0.05, ##P < 0.001, ###P < 0.0001 versus No inh, same time point (two-way ANOVA). e, Biosynthesis of 9-PA-D20HSA, 9-OA-D20HSA and 9-C17:1-D20HSA in HEK293T cells transfected with GFP, WT ATGL and ATGL(S47A) mutant enzymes. n = 4, mean ± s.e.m. *P < 0.01, **P < 0.001 versus GFP-transfected cells (one-way ANOVA). f–h, Biosynthesis of 9-PAHSA, 9-POHSA, 9-OAHSA and 9-C17:1HSA (f), biosynthesis of TG-esterified 9-FAHFAs (g) and levels of FFA and 9-HSA (h) in 3T3-L1 adipocytes incubated with C17:1 or 9-HSA alone or with or without atglistatin (10 µM). n = 3; mean ± s.e.m. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 versus vehicle, #P < 0.05, ##P < 0.01, ####P < 0.0001 versus C17:1 + 9-HSA group (one-way ANOVA). i–m, Biosynthesis of 9-PA-D20HSA, 9-PO-D20HSA and 9-OA-D20HSA (i), biosynthesis of 9-C17:1-D20HSA (j), biosynthesis of TG-esterified 9-FA-D20HSA (k), levels of TG-esterified C17:1 and D20HSA (l) and levels of FFA and HSA (m) in AT-Atgl-KO and Atglfl/fl SVF adipocytes incubated with atglistatin (10 µM) or 0.1% DMSO (No inh). TG-esterified 9-FA-D20HSA levels were measured in n = 4 samples per condition. FFA and HSA levels were measured in the Veh condition; n = 4 mean ± s.e.m. *P < 0.05, **P < 0.0001 versus WT, same treatment, #P < 0.0001 versus No inh, same genotype (two-way ANOVA, i, j, t-test two-tailed, k). Levels above the limit of quantification are shown with white circles. Similar results were obtained in two independent experiments.