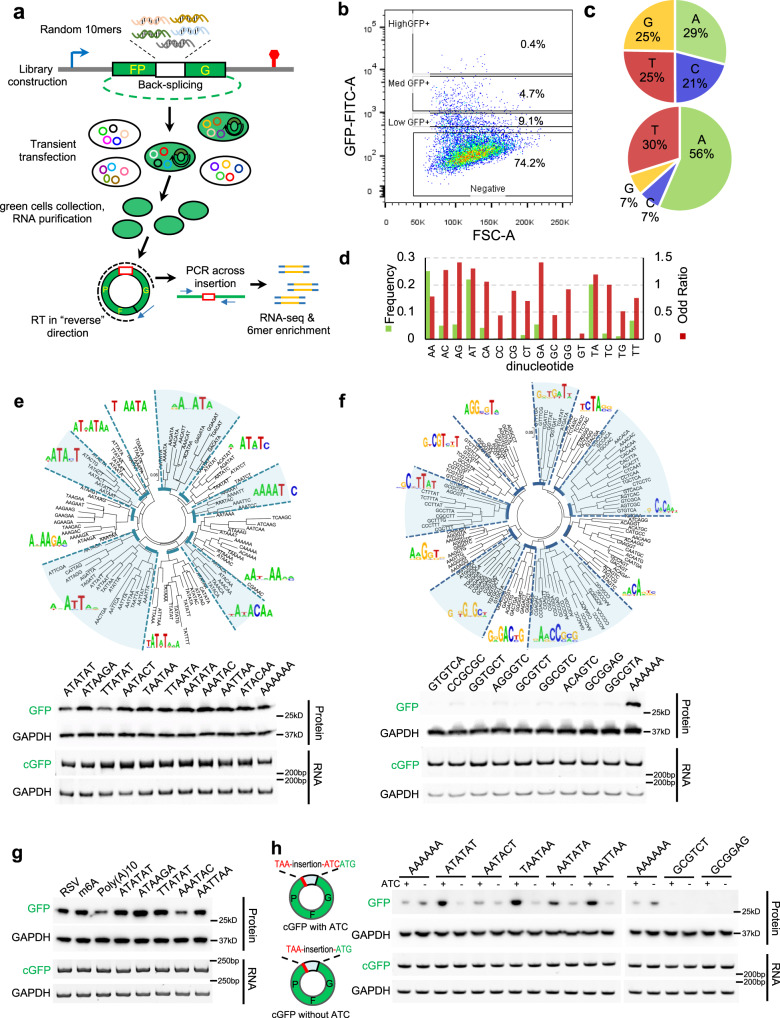
Fig. 1. Extensive IRES-like elements can drive circRNA translation.
Source data are provided in a Source Data file for panels (e–h). a Random decamers were inserted into pcircGFP-BsmBI, and the resulting library was transfected into 293T cells and the green cells sorted by FACS. The inserted sequences were recovered with RT-PCR and sequencing. The primers for RNA-seq library production were indicated by blue arrows. The enriched hexamers were identified computationally. b Flow-cytometry of cells transfected with circRNA reporter containing the 10-mer library. The cells were classified into four groups based on their GFP fluorescence (GFP negative, low, medium, or high GFP cells). The cells with medium and high fluorescence were sorted as “green cells”. c Single nucleotide frequency in the starting library (top) and the sequences enriched in green cells (bottom). d Frequencies and odd ratios of the dinucleotide in the sequences enriched in green cells. The odd ratio is defined as the probability of a dinucleotide divided by the product of the probabilities of each base. e The 97 enriched hexamers (i.e., IRES-like elements, z score >7) were clustered into 11 groups with the consensus motifs shown as pictogram (top). Representative hexamers in each cluster were inserted back into the circRNA reporter, which were transiently transfected into 293T cells. The GFP was assayed by western blot at 48 h after transfection (bottom). f The 122 depleted hexamers (i.e., negative control, z score < -7) were clustered into 11 groups and the consensus motifs were shown as pictogram (top). Representative hexamers in all clusters were tested using the same condition as panel (e), with AAAAAA as the positive control. g Comparison of newly identified IRES-like elements with m6A sites (RSV and m6A) for the activity to drive circRNA translation using the same condition as panel (e). h Effects of neighboring sequence on circRNA translation. The enriched and depleted hexamers were inserted into circRNA reporters with or without ATC trimer (partially resemble Kozak sequence). The samples were analyzed using the same condition as panel (e). The circRNA reporters inserted with poly-A sequence were loaded twice as control.