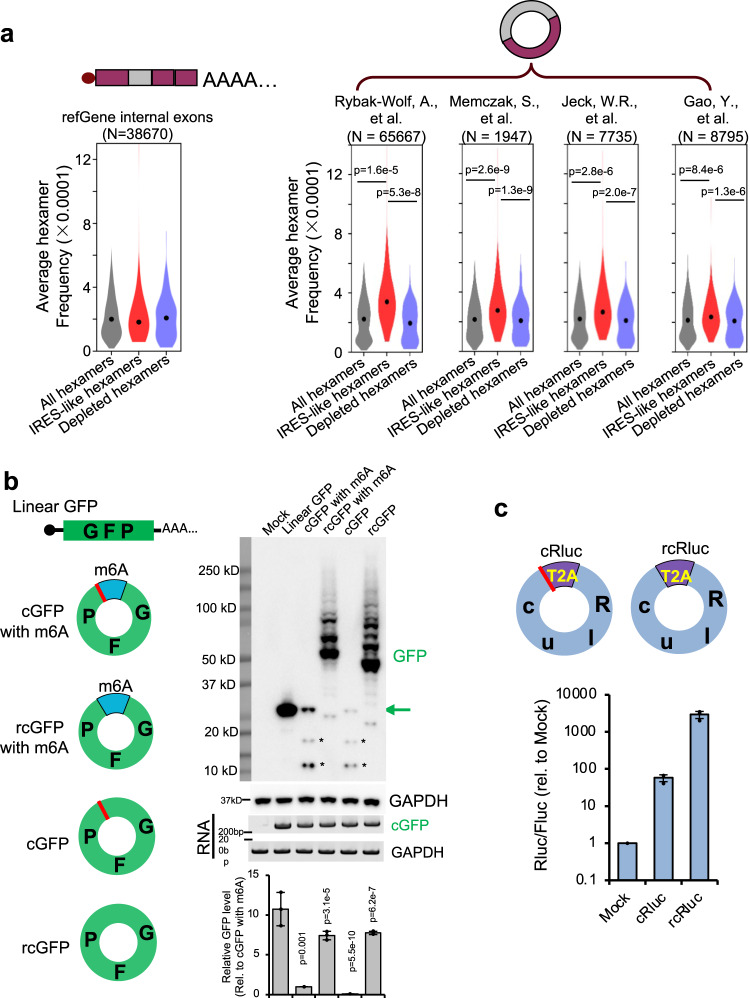
Fig. 2. CircRNAs contain many IRES-like elements that initiate translation.
Source data are provided as a Source Data file for panels (b) and (c). a IRES-like elements are significantly enriched in circRNAs. Average frequencies of different types of hexamers (all hexamers, IRES-like hexamers and depleted hexamers) in the internal exons of linear mRNAs vs. circRNAs were plotted. N: the number of RNA sequences in each dataset. The p-values were calculated with two-sided Kolmogorov–Smirnov test. b Translation of circRNAs can be initiated by internal coding sequence. Left panel, schematic diagrams of expression reporters for a linear mRNA and four circRNAs that code for GFP. From top to bottom: linear GFP mRNA; circRNA with an m6A site at upstream of the start codon; circRNA with an upstream m6A site and no stop codon; circRNA with start codon immediately following the stop codon; circRNA containing only the coding sequence (no stop codon or UTR). Red line, stop codon. The circRNA plasmids were transfected into 293T cells, and samples were analyzed by western blot at 2 days after transfection (right panel). The level of GAPDH was measured as a loading control. The green arrow indicates the full-length GFP, and the asterisks indicate the truncated GFP proteins translated using internal start codons. The bar graph represents the quantification of GFP protein levels relative to GAPDH. The protein levels were also normalized to the RNA. c Translation of the circRNA-coded Renilla luciferase (Rluc) using internal IRES-like elements. Top: schematic diagram of two Rluc circRNAs, where the Rluc ORF was split into two parts so that the full-length Rluc ORF can only be generated through back-splicing. The cRluc contains a sequence coding a T2A peptide by which the translation product can be cleaved into full length Rluc protein. rcRluc does not contain stop codon. Bottom: dual luciferase assay of cRluc and rcRluc. Control and circular Rluc plasmids were co-transfected with Fluc (firefly luciferase) reference reporter into 293T cells. The cells were lysed at 48 h after transfection for luminescence measurement using luminescence reader, and the relative luminescence signals were plotted.