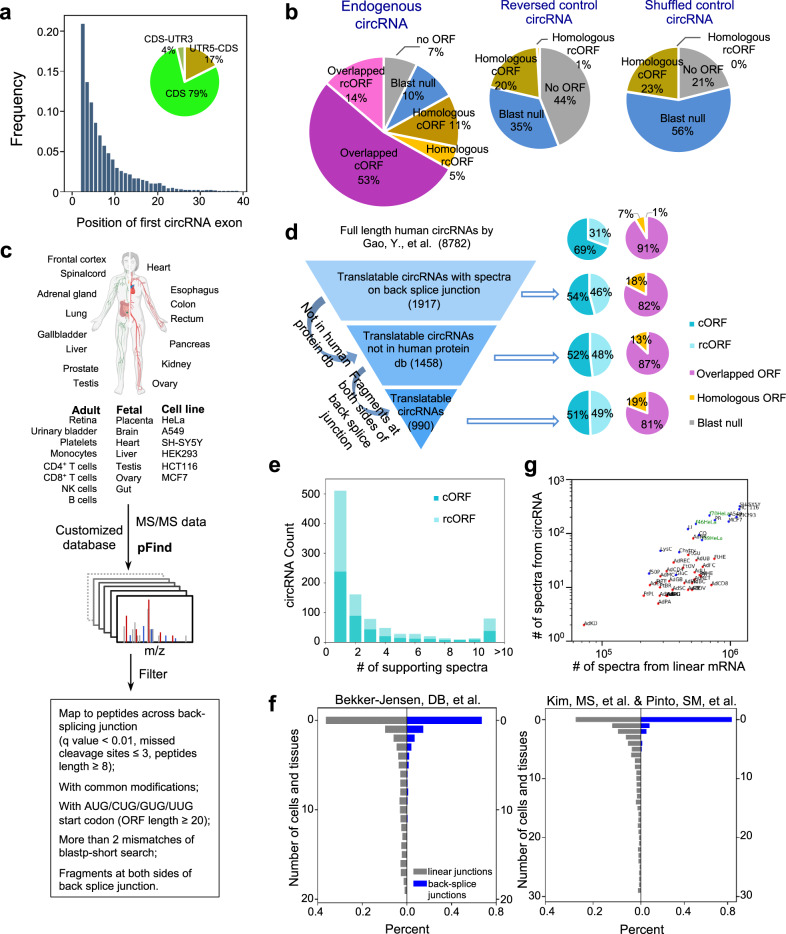
Fig. 4. Identification of circRNA-coded proteins.
a Position distribution for the first circRNA exon in their host genes. Full-length circRNA sequences were analyzed based on previous datasets, and the histogram was plotted according to the exon number of the first circRNA exon in host genes. The inserted pie chart presents the percent of circRNAs overlapping with different mRNA regions. b Survey of potential circRNA-coded products. Left, percent of endogenous circRNAs with an ORF > 20 aa. Purple pie slices: circRNAs translated regularly (cORF, dark purple) or in a rolling circle fashion (rcORF, light purple) into proteins that are partially overlapped with their host genes for at least 7aa; brown slices: circRNAs translated in a regular fashion (cORF, dark brown) or a rolling circle fashion (rcORF, light brown) into proteins that are not overlapped with their host genes but are homologous to other known proteins; blue pie slices: circRNAs translated into proteins that are not homologous to any known protein (cORF and rcORF combined); gray slices: circRNAs do not contain any potential ORF longer than 20 aa. Right panels: the same analysis of putative coding products from two control circRNA sets: reversed sequences or randomly shuffled sequences of the endogenous circRNAs. c Schematic diagram to identify circRNA-coded proteins using proteomic datasets. d Left, computational filters sequentially applied to identify translatable circRNAs and the numbers of circRNAs passing each filter. Right, the percentage of different types of circRNA-coded ORFs in the circRNAs passing each filter. The definition of different circRNA-coded ORFs is the same as panel (b). e Distribution of the supporting spectra for each translatable circRNA. f Distribution of the number of cell lines and tissues for each translatable circRNA in two proteomic datasets. g Comparison of the numbers of spectra from linear mRNAs vs. circRNAs. Abbreviations of different tissues and cell lines are listed in Supplementary Data 4. The proteomic data from Bekker-Jensen, DB, et al (blue) and Kim, MS, et al. & Pinto, SM, et al (red) were used. Green words indicate the 39 fractions, 46 fractions and 70 fractions from HeLa cells using high-capacity offline HpH reversed-phase LC.