Figure 1.
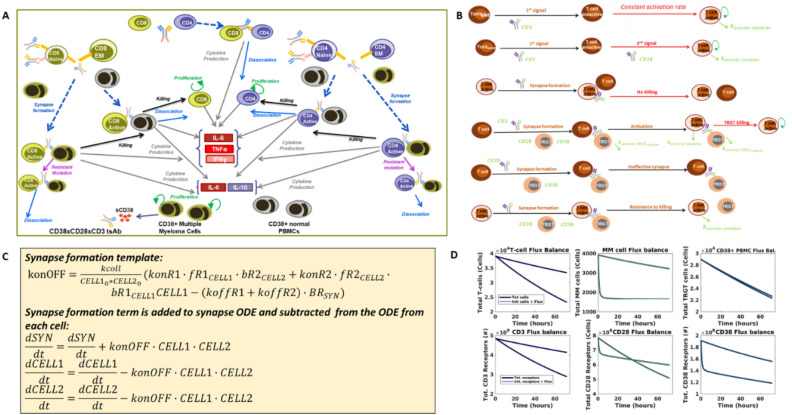
Our trispecific T-cell engager model was designed through a rigorous development process. Here we depict the four key steps in our model development process. (A) We first formulated a model diagram showing all the cells and interactions to be included in the model. We included eight key cells—naïve, effector memory, and active T-cells of CD4 or CD8 lineage, multiple myeloma cells, and CD38 + PBMCs. Interactions and processes included in the model are shown with arrows and labeled. (B) Assumptions made to determine how interactions should be mathematically formulated in the model are shown. Assumptions were made about the process of activation, synapse formation, killing, and resistance to killing based on literature research and internal discussions (Table S1). (C) The model code was generating by encapsulating all cells, synapses, receptors, and interactions in a rule-based model generation code. An example of the template used for the synapse formation term is shown. This term is added to the ODE for the synapse and subtracted from ODEs of the cells joining the synapse. (D) We verified that the assumptions and ODEs generated were properly executed by ensuring that flux balances were conserved in the model. The rule-based code produced an ODE for the net flux in each cell and receptor type (i.e. amount added/subtracted over time), which was then added to the initial cell/receptor number (blue lines). We compared this to the total number of cells and receptors across the simulation (black lines) to ensure that all species are conserved. In each panel, results are shown for two doses (8.4e−4 nM and 0.672 nM).