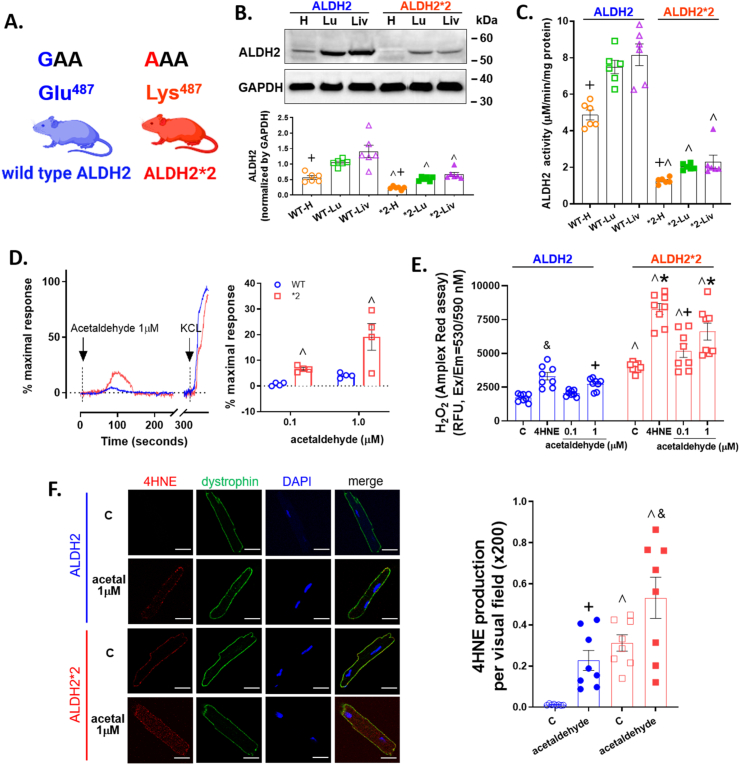
Fig. 2.
Basal level of ALDH2 expression and activity and cardiac myocyte response to acetaldehyde. A. An ALDH2*2 knock-in rodent on a C57/BL6J background was generated with a missense mutation (glutamic acid to lysine at 487) that reflects the human ALDH2*2 genetic variant. B. Top: Representative Western blot of ALDH2 protein (56 kDa) expression for heart, lung, and liver tissue homogenates for ALDH2 and ALDH2*2 mice. Bottom: Quantification of ALDH2 protein expression normalized to GAPDH (36 kDa) (n=6/group). C. ALDH2 enzymatic activity for homogenates of each genotype and organ (n=6/group). D. Calcium influx by 1 μM acetaldehyde. KCl (60 mM) was used as a positive control. E. H2O2 activity by 0.1 or 1 μM acetaldehyde with 4-HNE (20μΜ) as a positive control assessed by Amplex Red (n=8/group). F. Immunostaining of primary cardiomyocytes for 4-hydroxynoneal (4-HNE) protein adducts in vehicle or 1 μM acetaldehyde treated cells (left panel, bar = 20 μm). Green: dystrophin, Red: 4-HNE protein adducts, Blue: DAPI. Right: Quantification of 4-HNE protein adducts (n=8/group). WT= wild type, *2 = ALDH2*2, H= heart, Lu= lung, Liv= liver. Blue represents wild type ALDH2 mice and red represents the ALDH2*2 mice. ^p < 0.05, between genotypes; and +p < 0.05, &p < 0.01, *p < 0.001, within genotypes, respectively, calculated by one-way ANOVA with Bonferroni correction. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)