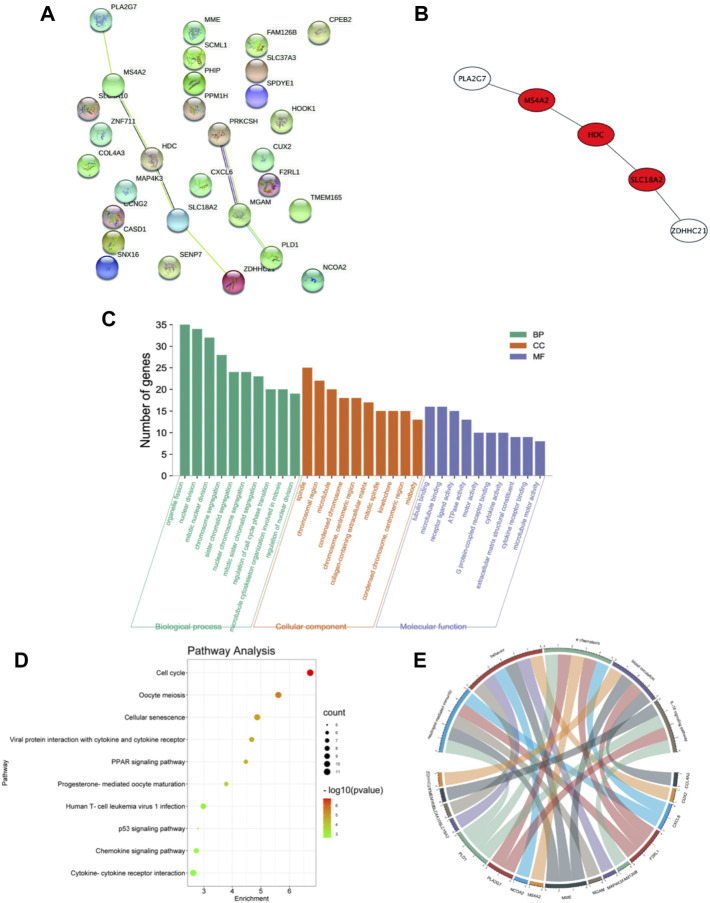
FIGURE 5.
PPI network and functional enrichment analysis of hub genes. (A) PPI network of 30 hub genes generated by the Cytoscape software. STRING was applied to create PPI network to define protein interactions. (B) The top 3 hub genes with the most correlations identified using CytoHubba, and the core gene scores were calculated using the Maximal Clique Centrality (MCC) method. (C) GO enrichment analysis results for hub genes. The functional enrichment analysis was performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID). GO terms enrichment for hub genes are shown in biological processes, molecular function, and cellular components. Y-axis shows the number of genes, and the x-axis shows the terms of the GO pathway. (D) The Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment: The sizes of the circle dots indicate the numbers of enriched genes, and the color of circle color indicates the p-value. The x-axis represents the number of genes in enrichments. (E) The GO-enriched chord diagram shows the genes involved in the GO term.