Figure 3.
Quantification of proviruses and multiply spliced HIV transcripts in FACS-sorted CD4+ T cells
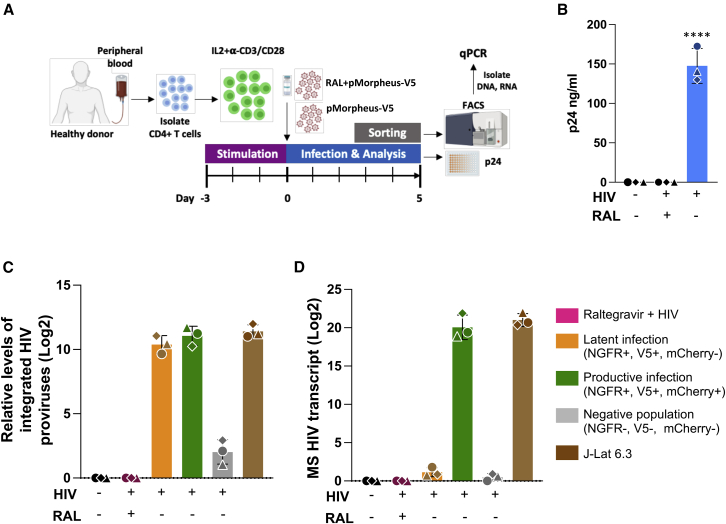
(A) Representation of the experimental design to analyze provirus integration or multiply spliced transcripts in productive or latently infected cells from three different healthy donors. CD4+ T cells were activated with IL-2 and α-CD3/CD28 antibody-coated beads for 3 days prior to infection with pMorpheus-V5. CD4+ T cells were infected with pMorpheus-V5 in the presence and absence of raltegravir. Latently (orange), productively (green), and the reporter negative cell populations were sorted using FACS 5 dpi with pMorpheus-V5. RNA and DNA were extracted from the same samples.
(B) Gag p24 concentration of culture supernatants was measured using ELISA at 5 dpi. Bar graph shows the production of p24 in pMorpheus-V5 or mock-infected cells in the presence and absence of raltegravir (RAL). Each donor is identified by a specific symbol. The average of three donors is shown (±SD) (∗∗∗∗p < 0.0001, unpaired t test).
(C) HIV integration was measured using a two-step quantitative Alu-PCR assay using total DNA from each sorted cell population (latent infection, yellow; productive infection, green; reporter expression-negative cells, gray). The bar graph depicts relative amounts of integrated proviruses. J-Lat 6.3 cells were used as a positive control. The mean ± SD of three independent experiments/donors are shown. Sorted cells from the same donor (N = 3) are identified by the same symbol.
(D) Multiply spliced (MS) HIV transcripts were quantified using semi-nested real-time qPCR. RNA was extracted from the same samples as in (C). RNA extracted from stimulated J-Lat 6.3 cells was used as a positive control. The bar graph depicts the mean ± SD from three independent experiments/donors. Sorted cells from the same healthy donor (N = 3) are identified by the same symbol.