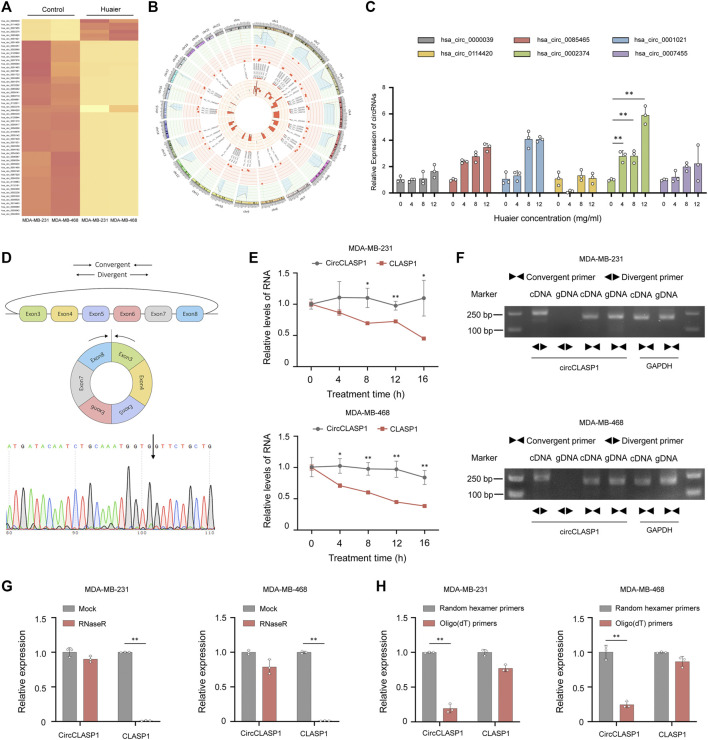
FIGURE 5.
CircCLASP1 is a candidate regulator of Huaier-induced ICD. (A) The heatmap showing the up- and down-regulated circRNAs in TNBC cells. (B) The differentially expressed circRNAs on human chromosome were visualized in the circos plot. The outermost circle showed the chromosomal distribution of candidate circRNAs. The second circle presented the p values. The third circle demonstrated the fold change between two groups. The fourth circle was used to exhibit the circBase IDs of selected circRNAs, and the innermost circle indicated the circRNA expression levels. (C) Validation of up-regulated circRNAs in Huaier-treated MDA-MB-231 cells by qRT-PCR. (D) CircCLASP1 is derived from exon 3 to 8 of CLASP1 gene. The backsplice junction site of circCLASP1 was identified by Sanger sequencing (black arrow). (E) qRT-PCR analysis of circCLASP1 and CLASP1 in Actinomycin D treated MDA-MB-231 and MDA-MB-468 cells. (F) CircCLASP1 could be amplified by divergent primers in cDNA but not gDNA. CLASP1 could be amplified by convergent primers in both cDNA and gDNA. GAPDH was used as a linear control. (G) The RNA levels of circCLASP1 and liner CLASP1 was detected by qRT-PCR in MDA-MB-231 and MDA-MB-468 cells treated with or without RNase R. (H) qRT-PCR analysis of relative expression levels of circCLASP1 and CLASP1 using random hexamer primers and oligo (dT) primers, respectively. *p < 0.05; **p < 0.01.